Extracellular vesicles as a promising biomarker resource in liquid biopsy for cancer
Abstract
Liquid biopsy is a minimally invasive biopsy method that uses molecules in body fluids as biomarkers, and it has attracted attention as a new cancer therapy tool. Liquid biopsy has considerable clinical application potential, such as in early diagnosis, pathological condition monitoring, and tailored treatment development based on cancer biology and the predicted treatment response of individual patients. Extracellular vesicles (EVs) are lipid membranous vesicles released from almost all cell types, and they represent a novel liquid biopsy resource. EVs carry complex molecular cargoes, such as proteins, RNAs [e.g., mRNA and noncoding RNAs (microRNA, transfer RNA, circular RNA and long noncoding RNA)], and DNA fragments; these cargoes are delivered to recipient cells and serve as a cell-to-cell communication system. The molecular contents of EVs largely reflect the cell of origin and thus show cell-type specificity. In particular, cancer-derived EVs contain cancer-specific molecules expressed in parental cancer cells. Therefore, analysis of cancer-derived EVs might indicate the presence and nature of cancer. High-speed analytical technologies, such as mass spectrometry and high-throughput sequencing, have generated large data sets for EV cargoes that can be used to identify many candidate EV-associated biomarkers. Here, we will discuss the challenges and prospects of EV-based liquid biopsy compared to other biological resources (e.g., circulating tumor cells and cell-free DNA) and summarize the novel studies that have identified the remarkable potential of EVs as a cancer biomarker.
Keywords
INTRODUCTION
Our body fluids, such as blood, urine, cerebrospinal fluid, saliva, pleural effusion, ascites fluid, breast milk and seminal plasma, contain various biological molecules[1]. Liquid biopsy is a minimally invasive method that uses these molecules as biomarkers, and it is emerging as a new tool in the strategy against cancer[2]. The terms "precision medicine"[3] and "personalized medicine"[4] have recently become popularized in the field of cancer research. Diagnosis and adequate therapy for individual cases of particular cancer types commonly rely on genetic mutation and gene expression analyses or pathological imaging observations of cancer lesions. However, such cancer genomic medicine approaches require collection of cancer tissue by biopsy, which imposes a heavy burden on the patient. In particular, it is difficult to obtain tumor tissue from organs located in the deeper parts of the body. Therefore, the development of minimally invasive methods, such as liquid biopsy, is desired. Liquid biopsy initially emerged only for the purpose of genetic diagnosis; however, it has considerable clinical application potential, such as in early diagnosis, monitoring of pathological conditions, and tailored treatment development according to cancer biology and the predicted treatment response[5]. Importantly, because of its minimally invasive nature, liquid biopsy can be scheduled more often to give more accurate snapshots of the disease at successive time points, which is useful for measuring temporal tumor burden levels and early evidence of recurrence or therapy resistance[6] [Figure 1]. Furthermore, liquid biopsy may reflect the genetic profile of more cancer subclones in a patient than tissue biopsies, which are obtained from only one cancer region[7].
Figure 1. Clinical utility of liquid biopsy in cancer. Liquid biopsy presents a minimally invasive nature and thus has the potential to impact clinical practice at multiple stages of cancer management. This technique can contribute to early diagnosis, pathological condition monitoring, and tailored treatment development according to the cancer biology of individual patients. After cancer treatment, liquid biopsy can support follow-up care by providing early evidence of recurrence or therapy resistance.
Extracellular vesicles (EVs) have attracted increasing attention as a novel analyte in liquid biopsy[8]. EVs are lipid membranous vesicles that are released from almost all types of cells, including normal cells as well as abnormal cells, such as cancer cells. EVs are reported to be correlated with various biological phenomena and play important roles in cell-to-cell communication via horizontal transfer of cellular cargoes, such as proteins, RNAs [including mRNA and noncoding RNAs (e.g., microRNA; miRNA, transfer RNA; tRNA circular RNA; circRNA and long noncoding RNA; lncRNA)], DNA fragments, and lipids[9,10]. Importantly, the composition of EV cargoes secreted from individual cell types differs greatly depending on the cellular origin, and the characteristic features of EVs derived from various cancer cells have been revealed. Thus, cancer-derived EVs can be analyzed to determine the presence and nature of cancer. Furthermore, EVs have been reported to be found in almost all body fluids[11]. For these reasons, EVs are recognized as a promising liquid biopsy resource for cancer. In this review, we will discuss the feasibility and practicality of EV-based liquid biopsy in clinical settings. In the first half, we will argue the advantages and challenges of EV-based liquid biopsy for clinical application. In the second half, we will summarize recent notable studies investigating cancer-specific EV-related molecules as cancer biomarkers, with a focus on their biological or clinical significance.
CANDIDATE ANALYTES IN LIQUID BIOPSY OTHER THAN EVS
Liquid biopsy can target various cancer-associated analytes in multiple body fluids, and several candidate analytes in liquid biopsy for cancer have been identified. These analytes include circulating tumor cells (CTCs) and circulating nucleic acids, such as cell-free DNA (cfDNA) and some extracellular RNA (exRNA) fragments [Figure 2]. Currently, CTCs and cfDNA are the most widely studied target materials for liquid biopsy[12,13].
Figure 2. Candidate analytes of liquid biopsy. Body fluids contain several promising biomarkers for cancer. Each candidate analyte can provide considerable information about the cancer biology of individual patients.
CTCs are cancer cells shed by primary or metastatic cancer lesions into the circulation and are considered a crucial determinant of hematogenous metastasis and recurrence. CTCs contain valuable information about the spreading tumor, and early detection of CTCs and treatment of metastatic spread can contribute to improving disease outcomes. Numerous studies in the past decade have shown that CTCs have potential as biomarkers to predict cancer metastasis progression and prognosis[14,15]. A high number of CTCs has been reported to be correlated with clinical outcome[16-19]. However, CTCs are estimated to account for at most one cell among a hundred million circulating cells; thus, there are generally only a small number of CTCs in a few milliliters of blood sample. Furthermore, CTCs must be analyzed as soon as possible after collection because the number of viable cells decreases rapidly. Therefore, analysis of CTCs requires relatively large volumes of fresh blood and advanced technology with extremely high analytical sensitivity and specificity[20-22]. To date, numerous technologies are available that are useful for enrichment and detection of CTCs[23]. Among them, the CellSearch® system has received FDA approval for prognostic clinical evaluation of several cancer types[24]. However, due to the limited number of CTCs in the blood and the current level of detection technologies, CTCs have not yet been entirely accepted in the clinic. Research groups have recently moved towards analyzing CTC contents (e.g., miRNA[25], mRNA[26], and protein[27,28]) for detection of biomarkers.
cfDNA is a short fragment of nucleic acids found in body fluids, and a component of cfDNA derived from tumor cells is called circulating tumor DNA (ctDNA). The majority of ctDNA originates from apoptotic and necrotic tumor cells. Over the past decade, a large number of studies have reported that measuring ctDNA levels in cancer patients may help in cancer diagnosis and prognosis prediction[29-31]. Interestingly, ctDNA is reported to be horizontally transferred from cancer cells to normal cells via uptake of apoptotic bodies, which is one of the EV subclasses (described below), leading to cancer progression[32,33]. Furthermore, ctDNA is also reported to harbor genetic and epigenetic changes present in the original tumor, and analytical techniques to detect such changes have already been established. By examining these changes, researchers have shown that ctDNA also helps predict treatment response and recurrence[34-38]. The major challenge in ctDNA research is that tumor-specific mutations may only represent 0.01% of the total cfDNA[39,40], thereby increasing the difficulty of detecting rare variants. Moreover, issues limiting cfDNA testing include its relatively short half-life[41,42]. Therefore, sample processing times are critical, and sample preservation requires special precautions, which are also significant barriers to practical use of cfDNA testing in the clinic.
CTC and ctDNA applications are confronted with several challenges, as described above. In short, the significantly low amounts and fragility of CTCs and ctDNA, which show remarkable variations in amount among individuals, increase the difficulty of detection. Moreover, CTC and ctDNA applications may be confined to evaluation of advanced cancer because CTCs are released from cancer tissue that has grown to the point that it causes metastases and ctDNA is primarily released from apoptotic or necrotic cancer tissue[43]. Thus, these entities may not be suitable for early diagnosis and may not reflect the current or near-future state of the disease.
exRNA refers to RNA that is present outside of cells within EVs or associated with platelets, lipoproteins and protein complexes. Similar to cfDNA, almost all non-EV-associated exRNAs are released into circulation by passive secretion. In addition to CTCs and cfDNA, non-EV-associated exRNA has also been well investigated as a candidate analyte in liquid biopsy for cancer. In particular, non-EV-associated miRNAs are recognized as a significant RNA subtype for biomarker discovery because miRNA profiles are associated with cancer-specific conditions. The critical issue is miRNA extraction from biofluid samples; the amounts of miRNA are highly variable among different experiments. Because of the small size of miRNAs and their attachment to other molecules, reproducible extraction remains an inherent issue[44].
WHAT ARE “EVS”?
We will now describe EVs, which are the main focus of this review. “EV” is a collective term that refers to all types of lipid membranous vesicles naturally released from all cell types. From the 1970s to the 1980s, membrane-enclosed vesicle structures were reported in various types of solid tissues, body fluids, and culture supernatants. Depending on their size and origin, these vesicles are called various names, such as prostasomes, exosomes, microvesicles, microparticles, shedding vesicles, ectosomes, apoptotic bodies and oncosomes. Each vesicle type contains slightly different molecular groups due to differences in vesicle biogenesis and secretion pathways. Nevertheless, their designations have been vaguely defined. Therefore, the International Society for Extracellular Vesicles (ISEV), founded in Sweden in 2012, recommends the use of “extracellular vesicle” as a generic term for these vesicles[45,46]. The nomenclature of these vesicles is detailed in the Minimal Information for Studies of Extracellular Vesicles (MISEV), which is the guideline advocated by the ISEV[47,48].
EVs are classically classified into three main categories: exosomes (approximately 100 nm), microvesicles (approximately 1 µm), and apoptotic bodies (greater than 1 µm)[49]. These three classes of EVs differ in size as well as morphology, content, generation mode, and release mechanism. Exosomes are formed by the inward budding of early endosomes to form multivesicular bodies (MVBs). These MVBs fuse with the limiting plasma membrane to release exosomes into the extracellular space. Microvesicles originate by direct shedding or budding from the plasma membrane. Apoptotic bodies are released from cells undergoing programmed cell death[50]. Recently, Théry et al[47,51]. developed a more reasonable classification system for EVs and defined vesicles < 100 nm as small EVs, < 200 nm as medium EVs, and > 200 nm as large EVs. Moreover, they demonstrated that each EV subtype showed different characteristic protein components, suggesting that each EV category is generated and secreted through a specific molecular mechanism[47,51].
EVS AS AN ANALYTE IN LIQUID BIOPSY
In 1983, Pan and Johnstone[52] discovered that cells secrete 100 nm-sized vesicles with a lipid membranous structure and that red blood cells release small vesicles loaded with the transferrin receptor, which is necessary to synthesize hemoglobin during maturation. They regarded EVs as “garbage bags” that are used to expel unused molecules from the cell and reported one of the remarkable EV functions; more importantly, this study demonstrated the presence of proteins in EVs[52,53].
In the 1990s, by analyzing immune cells, Raposo et al.[54] found that EVs affect recipient cells. EVs derived from B lymphocytes induced antigen-specific MHC class II-restricted T cell responses, indicating that EVs have functions related to intercellular communication[54].
In 2007, Valadi et al.[55] demonstrated that EVs from the human mast cell line HMC-1 and the mouse mast cell line MC/9 contain approximately 1300 mRNAs and 121 miRNAs and thus contribute to the exchange of genetic information between cells. Since that time, miRNAs have been the most widely studied class of short noncoding RNAs (ncRNAs) in EV cargoes[55].
To date, EVs have been reported to contain various proteins, RNAs [including mRNAs and noncoding RNAs (miRNAs, tRNAs, circRNAs and lncRNAs)], and DNA fragments[9]. The past few years have seen extraordinary developments in areas such as mass spectrometry (MS), high-throughput sequencing (HTS) (e.g., DNA sequencing, chromatin immunoprecipitation sequencing, methylation sequencing, and RNA sequencing), and big-data analysis[56,57]. These advanced technologies allow us to select analytes contained in EVs and evaluate them deeply. Significantly, the composition of EV molecular contents reflects the intracellular status of their cellular origin. Moreover, these molecules are stably preserved due to their lipid-layered structure; hence, EVs from body fluids can be analyzed after being stored for a relatively long period of time[58]. A growing body of evidence has suggested that cancer-derived EV cargoes bare a strong resemblance to the intracellular status of their parental cells[59]. Analysis of EV cargoes can help reveal the existence, molecular profile and behavior of cancer. Thus, EV-based liquid biopsy contributes to early cancer diagnosis, monitoring of cancer pathological conditions, and treatment selection according to cancer biology and the predicted treatment response. In addition, EVs are easily accessible in various kinds of body fluids and appropriate for sequential collection. For these reasons, EVs have emerged as a promising biomarker resource and as another kind of liquid biopsy for cancer [Figure 2].
CLASSICAL EV ISOLATION METHODS
Although EVs are a remarkable candidate analyte to detect cancer biomarkers, few EV-associated biomarkers have been implemented in clinical settings, which is partially due to the lack of adequate isolation methods[60]. In principle, EVs must be isolated from various biofluids for analysis. Indeed, in almost all cases, candidate EV-associated biomarkers were detected via isolation processes because target EV-associated molecules should be distinguished from EV-free molecules in the biofluid; however, a unified isolation method is not currently available, which is one of the biggest challenges in EV research.
The most popular technique for EV isolation is ultracentrifugation (UC), which separates EVs based on their size and buoyant density. The UC procedure does not require additional chemicals or pretreatment of the body fluid samples. Moreover, the combination of UC with other procedures, such as ultrafiltration, sucrose cushion, and density gradient centrifugation, can increase the purity of the EV fraction[61-63]. However, UC-based procedures generally require dedicated equipment and an extraordinary amount of time[60]. Moreover, repeated centrifugation can lead to reduced yield due to lost and aggregated EVs[64]. On the other hand, various polymer-based isolation kits, such as ExoQuick™, Total Exosome Isolation™ and miRCURY™, which are commercially available, can save time and labor costs, although issues with high contamination rates and high running costs have been reported[63,65]. An immunoaffinity capture approach using magnetic beads and monoclonal antibodies targeting EV surface antigens is also commonly used to isolate EVs[66,67]. Many reports have demonstrated that this approach achieves a selective and high-purity output; however, there are difficulties in terms of low capacity, low yield and high reagent cost[60]. Size-exclusion chromatography (SEC) is a technique for separating biological molecules based on molecular size in which target molecules are isolated by filtration through a resin-packed column. SEC has been reported to yield highly purified EVs and achieve excellent reproducibility in a relatively short time[60,64]; however, the existence of EVs in multiple fractions results in a low EV concentration in the obtained samples. Consequently, subsequent EV analysis often requires an additional concentration step[68]. Recently, many researchers have studied combined methods integrating multiple isolation techniques together or conducting them sequentially[69]. Among others, the combination of UC and SEC has demonstrated higher purity of EVs and better performance in subsequent analyses than the single-step methods[70-72]; however, in daily clinical settings, combined methods might be avoided due to several factors, such as time consumption, cost, repeatability, and ease of use. As high-quality EV samples are essential for subsequent analyses, there is an urgent need for a simple, cost-effective, and reliable technique for both basic research and clinical practice.
EV-ASSOCIATED PROTEINS
Common EV protein markers
EVs contain abundant proteins that reflect their origin and alterations of their parental cells. Based on the endosome-based biogenesis pathway, EV-specific protein markers include membrane trafficking-associated proteins (e.g., Rab family proteins, annexins), MVB-associated proteins (e.g., Alix, Tsg101 and ESCRT complex), tetraspanins (e.g., CD9, CD63 and CD81) and heat shock proteins (Hsp70 and Hsp90)[73-75]. As these proteins are common to EVs derived from almost all cell types, they can serve as common positive EV markers that can confirm the presence of EVs, and isolated EVs can be assessed via downstream proteomic analyses, such as western blotting (WB), enzyme-linked immunosorbent assay (ELISA), and flow cytometry (FCM), which use common EV proteins as hallmarks. With regard to checking the quality of isolated EV samples, MISEV recommends the use of apolipoproteins A1/2 and B and albumin as negative markers of blood-derived EVs because they are often co-isolated with these molecules[47,76].
EV protein-based platform for cancer diagnosis
Targeting membrane proteins on the surface of EVs is an effective strategy because target cancer-derived proteins can be directly detected without the use of a large sample volume or time-consuming isolation processes for EVs. Recently, great efforts have been devoted to establishing clinically useful detection platforms, including platforms for direct detection of cancer-specific EVs without any isolation or purification procedures. These platforms mainly consist of specific antibody-based technologies that detect EV surface proteins, such as ELISA.
Jørgensen et al.[77] established the EV Array, which can detect EVs in unpurified materials in a high-throughput manner. The EV Array is composed of different capture antibodies located on a microarray slide, which capture EVs according to their surface proteins, and the target EVs are detected with a cocktail of biotinylated antibodies against the tetraspanins CD9, CD63, and CD81. The authors validated the performance of the EV Array by comparing plasma from nonsmall cell lung cancer (NSCLC) patients and normal healthy subjects[78]. Shao et al.[79] reported that a microfluidic chip platform could distinguish patients with glioblastoma multiforme (GBM) from normal healthy subjects. This microfluidic chip labeled with magnetic nanosensors quantifies an EV-specific protein marker (CD63) and glioblastoma-specific proteins, such as epidermal growth factor receptor (EGFR) and EGFR variant III, on the surface of EVs via micronuclear magnetic resonance (μNMR). In this study, the authors also indicated that GBM EVs reflect gene amplification or mutation and predict the therapy response. Surface plasmon resonance (SPR)-based nanosensors have recently attracted much attention due to their ability to detect a small number of molecules[80]. Im et al.[81] developed an SPR-based exosome sensor called nanoplasmonic exosomes (nPLEXs). Each nanohole array of nPLEX is functionalized with antibodies that recognize EV surface proteins. nPLEX was able to differentiate ascites samples from ovarian cancer patients from healthy controls with an accuracy of 97% and identified ovarian cancer cell-derived EVs based on their expression of CD24 and EpCAM. Yoshioka et al.[82] also established a highly rapid and sensitive analytical technique called “ExoScreen”. This assay consists of two kinds of antibodies against proteins on the surface of EVs that are detectable by photosensitizing beads. A very small sample volume (at least 5 μL) was required to detect EVs in serum from healthy controls without a complicated isolation process. Moreover, the assay could be completed within 2 h. In this study, they identified CD147 as a specific EV-surface protein derived from colorectal cancer cells and revealed that a larger number of CD9/CD147 double-positive EVs could be detected in serum from colorectal cancer patients than in serum from healthy control subjects using this assay. Furthermore, Zhao et al.[83] developed a simple microfluidic platform named the “ExoSearch” chip that allows quantitative isolation of EVs using immunomagnetic beads. An “ExoSearch” chip could detect ovarian cancer by measuring three EV cancer protein markers, CA-125, EpCAM and CD24. The development of immune-capturing systems in microchips also provides highly sensitive and reliable detection of cancer markers without requiring a large sample volume or time-consuming EV isolation processes.
Other EV protein biomarkers for cancer diagnosis
EV surface proteins and EV lumen proteins are regarded as candidate cancer biomarkers. Numerous studies have identified cancer-associated EV protein markers using isolation processes, followed by WB, ELISA, and FCM. Promising EV protein markers identified in clinical studies with patient body fluid samples are summarized in Table 1.
A list of EV proteins as potential biomarkers for cancer
| Cancer types | Biological source | Isolation method | Detection method | Markers | Potential application | Ref. |
| Urinary cancer | ||||||
| Prostate cancer | Plasma | UC | ELISA | PSA | Diagnosis/Prognosis | [85] |
| Urine | UC + SUC | ELISA/WB | PSA, PSMA | Diagnosis/Monitoring | [84] | |
| Plasma/Serum | UC/ExoQuick | ELISA/WB | Survivin | Diagnosis/Monitoring | [87] | |
| Urine | UC | IP/WB | δ-catenin | Diagnosis | [156] | |
| Serum | UC | WB | MDR-1/P-gp, MDR-3, PABP4 | Predict chemoresistance (Docetaxel) | [95,157] | |
| Plasma | - | FCM | PSMA | Monitoring/Predict chemoresistance | [158] | |
| Bladder cancer | Urine | UC | MS/WB | EH-domain-containing protein 4, EPS8L1, EPS8L2, GTPase NRas, Mucin 4, retinoic acid-induced protein3, resistin, alpha subunit of GsGTP binding protein | Diagnosis | [159] |
| Urine | UC | ELISA | TACSTD2 | Diagnosis | [160] | |
| Urine | UC | MS | α-1-anti-trypsin, H2B1K | Diagnosis | [161] | |
| Urine | UC | WB | HEXB, S100A4, SND1, TALDO1, and EHD4 | Diagnosis | [162] | |
| Urine | UC | WB | Periostin | Diagnosis | [163] | |
| Urine | UC + SUC | WB | EDIL3 | Diagnosis | [164] | |
| Urine | UC + SUC | FCM | CD36, CD44, 5T4, basigin, CD73, MUC1, α6-integrin | Diagnosis | [165] | |
| Renal cancer | Urine | UC | MS/WB | MMP9, DKK4, EMMPRIN, CP, PODXL, CAIX, CD10, AQP1, dipeptidase 1, syntenin 1 | Diagnosis | [166] |
| Female cancer | ||||||
| Breast cancer | Serum | ExoQuick | ELISA | Survivin, Survivin2B | Diagnosis/Prognosis | [88] |
| Serum/Plasma | ExoQuick | ELISA/WB | CD82 | Diagnosis | [167] | |
| Ascites | UC + SUC | WB | CD24, EpCAM | Diagnosis | [168] | |
| Plasma | UC | FCM | TRPC5 | Prognosis/Predict chemoresistance | [169] | |
| Serum | UC | FCM | UCH-L1 | Predict chemoresistance (Anthracycline/taxan) | [92] | |
| Serum | UC | FCM/WB | HER2 | Predict chemoresistance (Trastuzumab) | [96] | |
| Plasma | UC | ELISA/FCM | TGFβ1 | Predict chemoresistance (Trastuzumab) | [97] | |
| Ovarian cancer | Plasma | - | Exosearch chip | CD24, EpCAM, CA-125 | Diagnosis | [83] |
| Plasma | UC | WB | TGFβ1, MAGE3/6 | Diagnosis | [170] | |
| Plasma | UC + SUC | WB | Claudin-4 | Diagnosis | [171] | |
| Ascites | UC + SUC | WB | E-cadherin | Diagnosis/Prognosis | [172] | |
| Ascites | UC | WB | MMP2, MMP9, uPA | Diagnosis | [173] | |
| Ascites | UC + SUC | WB | CD24, L1CAM, ADAM10, EMMPRIN | Diagnosis/Prognosis | [174] | |
| Digestive cancer | ||||||
| Pancreatic cancer | Serum | UC | FCM | GPC1 | Diagnosis/Prognosis | [89,175] |
| Serum | UC + SUC | ELISA | CKAP4 | Diagnosis | [176] | |
| Serum | UC | ELISA | MIF | Diagnosis/Prognosis | [177] | |
| Plasma | - | ELISA | EpCAM | Prognosis | [91] | |
| Colorectal cancer | Serum | Exoquick | ELISA | CEA | Diagnosis | [178] |
| Serum | - | ExoScreen | CD147, CD9 | Diagnosis | [82] | |
| Ascites | UC | WB | claudin-3 | Diagnosis | [179] | |
| Gastric cancer | Serum | UC | FCM/WB | HER-2/neu, CCR6, EMMPRIN, MAGE-1, c-MET | Diagnosis | [180] |
| Others | ||||||
| Lung cancer | Serum | UC | ELISA | EGFR | Diagnosis | [181] |
| (NSCLC) | Serum | UC | ELISA/WB | AHSG, ECM1 (with serum CEA) | Diagnosis | [182] |
| Serum | - | EV array | 30 Proteins | Diagnosis | [78] | |
| Plasma | - | EV array | CD171, NY-ESO-1, PLAP, Flotilin1 | Diagnosis | [183] | |
| Urine | UC | WB | LRG1 | Diagnosis | [184] | |
| Melanoma | Plasma | UC | ELISA (Exo Test)/FCM/WB | Caveolin-1, CD63 | Diagnosis/Prognosis | [185] |
| Plasma | UC | WB | TYRP2, VLA-4, HSP70, HSP90 | Prognosis | [186] | |
| Plasma | UC/Total Exosome isolation Kit | ELISA/FCM /WB | PDL-1 | Predict immunotherapy resistance (Pembrolizumab) | [91] | |
| Glioblastoma | Serum | - | µNMR system | EGFR, EGFRvⅢ, CD63 | Prognosis | [79] |
Clinically validated traditional molecules have also been identified in EVs, such as prostate-specific antigen (PSA), and they represent novel diagnostic biomarkers. Mitchell et al.[84] demonstrated that the expression of PSA and PSMA in urinary EVs can act as treatment response markers in prostate cancer. Additionally, Logozzi et al.[85] showed increased PSA expression on EVs in vitro and in the plasma of prostate cancer patients. The authors emphasized the failure of current PSA testing in discriminating between benign prostatic hypertrophy and prostate cancer in terms of both overdiagnosis and overtreatment, which leads to patient suffering and public and private healthcare expenditures. Moreover, they claimed that EV PSA might resolve the problem associated with differences in PSA cutoff levels based on age, race and individual physiological condition. Similar to PSA, some traditional molecules in EVs have been discussed to have higher relevance to cancer than their total amount in body fluids, such as CEA for colon cancer[86].
Additionally, some EV-associated markers have been reported as diagnostic markers for multiple cancer types. Khan et al.[87] showed that Survivin, an inhibitor of apoptosis member, could be detected in plasma-derived EVs from both prostate cancer patients and healthy subjects; however, the relative amount of EV Survivin was remarkably higher in the plasma of prostate cancer patients. Their subsequent study showed that the EV Survivin and its alternative splice variants were also elevated in breast cancer patient plasma[88], which suggests that EV Survivin might be an important diagnostic marker common to several cancer types.
EV proteins can be used to monitor cancer progression and drug resistance
Biomarkers in EVs of several cancer types may be applied for cancer stratification because they change in response to anticancer therapy. Thus, EV proteins may be used as novel biomarkers to monitor cancer progression or to identify patients susceptible to anticancer drugs.
Melo et al.[89] reported that glypican-1 (GPC1) could be a specific marker of cancer-derived EVs and that the existence of GPC1-positive EVs in serum could differentiate patients with pancreatic ductal adenocarcinoma (PDAC) from patients with benign pancreas disease or from healthy control subjects with 100% sensitivity and specificity. The levels of GPC1-positive EVs were significantly decreased after surgical resection; moreover, they were related to overall survival (OS) and significantly higher in patients with distant metastasis than in patients with lymph node metastasis only or no metastases[89]. Although this conclusion is controversial, a replication study was performed to validate this finding, and a subsequent discussion was held in ISEV2017[90]. Similarly, Giampieri et al.[91] reported that a higher level of EpCAM-positive EVs before chemotherapy was correlated with shorter progression-free survival (PFS) and OS. In contrast, in this study, an increase in EpCAM-positive EV levels during treatment was correlated with better PFS in PDAC patients[91]. These studies demonstrated that EV-associated proteins could be biomarkers for monitoring tumor burden.
The development of chemoresistance is a persistent problem during cancer treatment. Various studies have reported cell-to-cell transfer of multidrug resistance (MDR) efflux pumps as EV cargoes from chemotherapy-resistant cells to chemotherapy-sensitive cancer cells. EVs from doxorubicin-resistant[92] or docetaxel-resistant[93] breast cancer cell lines transferred chemoresistance to recipient cancer cells through P-glycoprotein (P-gp) loaded onto EVs. Moreover, the same phenomenon in paclitaxel-resistant ovarian cancer cells was also caused by the transfer of functional P-gp mediated by EVs[94]. Docetaxel-resistant prostate cancer cells were also reported to proliferate through cell-to-cell transfer of EV P-gp. In this study, serum EVs from prostate cancer patients who were nonresponders to docetaxel therapy protected prostate cancer cells from the cytotoxicity of docetaxel[95]. These studies indicate that EV-P-gp might be a promising marker to predict chemotherapy resistance in several cancer types. Ubiquitin C-terminal hydrolase L1 (UCH-L1) has also been reported as an EV-based predictive biomarker of chemoresistance in breast cancer. UCH-L1 overexpression has been reported to induce upregulation of P-gp levels through the MAPK/ERK signaling pathway, thereby enhancing an MDR phenotype in breast cancer. Ning et al.[92] showed that higher UCH-L1 levels in circulating EVs are correlated with poorer response to adjuvant anthracycline/taxane-based chemotherapy. In addition, they demonstrated that UCH-L1-positive EVs derived from breast cancer cell lines could transfer chemoresistance to recipient cells in vitro, indicating that EVs might be a predictive biomarker of chemoresistance in breast cancer patients.
Interestingly, EV-mediated drug resistance has been reported to be relevant for molecular target drugs as well as chemotherapy drugs. Ciravolo et al.[96] reported that 73% of advanced-stage breast cancer patients had HER2-positive EVs in circulation, which hampered the corresponding therapeutic efficacy of trastuzumab monoclonal antibody. Importantly, this study demonstrated that the presence of HER2-positive EVs in the serum of breast cancer patients could be an indicator to predict a patient’s response to trastuzumab therapeutic regimens[96]. Martinez et al.[97] reported that EVs released from HER2 drug-resistant cells contain larger amounts of the immunosuppressive cytokine TGF-β1. Importantly, a recent neoadjuvant clinical trial by the same group that included trastuzumab and lapatinib further demonstrated that HER2-overexpressing breast cancer patients who were nonresponders to HER2 drug therapy had significantly higher amounts of TGF-β1 in circulating EVs than patients who did not respond to HER2 drug therapy. These results suggest that EV-TGF-β1 can be a biomarker for predictive response to this therapeutic regimen against breast cancer[97].
Recently, with the successful development of immune checkpoint inhibitors (ICIs), cancer immunotherapy has attracted worldwide attention as a new cancer treatment. EV proteins may contribute to identifying patients susceptible to ICIs. Tumor cells avoid immune recognition by upregulating the surface expression of programmed death-ligand 1 (PD-L1), which interacts with the programmed death-1 (PD-1) receptor on T cells to elicit the immune checkpoint response. Indeed, immunotherapy with anti-PD-1 antibodies has shown remarkable therapeutic effects against different tumor types[98]. However, for some patients, the therapeutic response has been reported to be rather poor[99,100]. To address this problem, Chen et al.[101] showed that specific EVs reduce the effectiveness of immunotherapy approaches in certain patients with melanoma. Remarkably, using human melanoma xenografts in nude mice, they showed that metastatic melanoma cell lines release EVs loaded with PD-L1 on their surface and that interferon-γ increases the expression of PD-L1 on these vesicles, which suppresses the function of CD8+ T cells, facilitating tumor growth. Importantly, these authors showed that the level of PD-L1-positive EVs differentiates responders from non-responders to anti-PD-1 therapy. More importantly, this study provides evidence supporting the application of PD-L1-positive EVs as a predictive biomarker for anti-PD-1 therapy in melanoma patients[101].
EV-ASSOCIATED NUCLEIC ACIDS
As described above, high-speed analytical technologies, such as HTS, are powerful tools for identifying candidate nucleic acid biomarkers. For detection and validation of candidate EV nucleic acid biomarkers, reverse transcription-quantitative polymerase chain reaction (RT-qPCR) after an isolation process is the most common detection method. Recently, digital polymerase chain reaction (dPCR) has emerged as a novel detection method for EV nucleic acids. dPCR is a qPCR technique that provides a sensitive and reproducible method of measuring the amount of DNA or RNA present in a sample[102]. The tremendous progress in these technologies during the last few decades has allowed us to analyze EV-associated nucleic acids derived from diverse body fluids.
EV RNA biomarkers
Pioneering studies of nucleic acids from isolated EVs have identified various miRNAs and mRNAs as the major components of EVs[55,103]. Subsequently, several important papers reporting on the function of EV-miRNAs were published and showed that the transferred EV-miRNAs can be active in recipient cells and modify the cellular phenotype[104-106]. Over the last decade, studies have revealed that EVs contain other noncoding RNAs, such as tRNAs[107], circRNAs[108] and lncRNAs[109,110]. To date, EV RNAs (as EV cargo) have received the most attention in terms of cancer diagnosis and prognosis because they are easy to quantify using conventional methods, such as qPCR, and stable against RNase-dependent degradation in the circulatory system[111]. Table 2 summarizes the candidate EV RNA markers reported as diagnostic and prognostic tools for cancer.
A list of EV RNAs as potential biomarkers for cancer
| Cancer types | Biological source | Isolation methods | Detection methods | RNA types | Markers | Potential application | Ref. |
| Uninary cancer | |||||||
| Prostate cancer | Urine | UC + SUC | nestedPCR | mRNA | PSA, PCA-3, TMPRSS:ERG | Diagnosis/Monitoring | [115] |
| Urine | - | ExoDx® Prostate (IntelliScore) | mRNA/lncRNA | PCA3, ERG, SPDEF | Diagnosis | [118] | |
| Urine | UF/UC | RT-qPCR | miRNA | let-7c, miR-21, miR-107, miR-145, miR-196a-5p, miR-204, miR-375, miR-501-3p, miR-574-3p, miR-2909 | Diagnosis | [116,187-191] | |
| Serum/Plasma | UF/SEC/UC | RT-qPCR | miRNA | let-7i, miR-16, miR-21-5p, miR-24, miR-26a, miR-26b, miR-30c-5p, miR-34b, miR-92b, miR-93, miR-103, miR-106a, miR-107, miR-130b, miR-141, miR-181a-2, miR-195, miR-197, miR-200c-3p, miR-210-3p, miR-223, miR-298, miR-301a, miR-326, miR-328, miR-331-3p, miR-346, miR-375, miR-432, miR-574-3p, miR-625, miR-1290, miR-2110 | Diagnosis | [187,192-196] | |
| Plasma | UF/SEC/UC | RT-qPCR | miRNA | miR-17, miR-20a, miR-23a, miR-130b, miR-198, miR-200b, miR-375, miR-379, miR-513a-5p, miR-572, miR-577, miR-582-3p, miR-609, miR-619, miR-624, miR-1236, miR-1290 | Prognosis | [119,187] | |
| Serum | UC | RT-qPCR | miRNA | miR-1246 | Prognosis | [197] | |
| Urine | - | Urine Exosome RNA Isolation Kit / RT-qPCR | lncRNA | p21 | Diagnosis | [122] | |
| Plasma | Total Exosome Isolation Kit | RT-qPCR | lncRNA | SAP30L- AS1, SChLAP1 | Diagnosis | [198] | |
| Plasma / Urine | - | exoRNeasy kit/ddPCR | mRNA | AR-V7 | Predict hormone therapy resistance (Abiraterone and/or Enzalutamide) | [132,133] | |
| Bladdar cancer | Urine | UC | ddPCR | miRNA | miR-21, miR-93, miR-200c, miR-940 | Diagnosis | [199] |
| Urine | - | ddPCR | miRNA | miR-21-5p, miR-4454, miR-720, miR-200c-3p, miR-29b-3p, miR-200b-3p | Diagnosis | [200] | |
| Urine | UC | RT-qPCR | miRNA | miR-375, miR-146a | Diagnosis | [201] | |
| Urine | unknown | RT-qPCR | miRNA | miR-146b-5p, miR-155-5p | Diagnosis | [202] | |
| Urine | UC | RT-qPCR | lncRNA | HOTAIR, HOX- AS-2, MALAT1, SOX2, OCT4 | Diagnosis/Prognosis | [203] | |
| Serum | ExoQuick | RT-qPCR | lncRNA | PCAT1, UBC1, SNHG16 | Diagnosis/Prognosis | [204] | |
| Serum | UC | RT-qPCR | lncRNA | UCA1 | Diagnosis | [205] | |
| Urine / Serum | UC | RT-qPCR | circRNA | PRMT5 | Diagnosis/Prognosis | [126] | |
| Renal cancer | Serum | UC + IP | RT-qPCR | miRNA | miR-210, miR-1233 | Diagnosis | [206,207] |
| Urine | UC | RT-qPCR | miRNA | miR-150-5p, miR-126-3p in combination with miR-34b-5p, miR-449a and miR-486-5p | Diagnosis | [208] | |
| Serum | Total Exosome Isolation Kit | RT-qPCR | miRNA | miR-224 | Prognosis | [209] | |
| Plasma | ExoQuick | RT-qPCR | miRNA | miR-26a-1-3p, miR- let-7-I, miRN-615-3p | Prognosis | [210] | |
| Urine | UC | RT-qPCR | lncRNA | GSTA1, CEBPA, PCBD1 | Diagnosis | [211] | |
| Female cancer | |||||||
| Breast cancer | Plasma | FC/UC/ExoQuick | RT-qPCR | miRNA | miR-21, miR-1246 | Diagnosis | [212] |
| Plasma | UC | RT-qPCR | miRNA | miR-105 | Diagnosis | [213] | |
| Serum | unknown/Total Exosome Isolation Kit | RT-qPCR | miRNA | miRNA-21, miRNA-222, miRNA-155 | Prognosis/Predict chemoresistance (miR-21, miR-155: Doxorubicin, Paclitaxel) | [214,215] | |
| Serum | UC + IP | RT-qPCR | miRNA | miR-200a, miR-200c, miR-205 | Diagnosis | [216] | |
| Bood/Milk/Ductal fluids | UC | RT-qPCR | miRNA | miR-16, miR-1246, miR-451, miR-205 | Diagnosis | [217] | |
| Serum | ExoQuick | RT-qPCR | lncRNA | HOTAIR | Prognossis/Monitoring | [129] | |
| Serum | ExoQuick | RT-qPCR | lncRNA | SNHG14 | Predict chemoresistance (Trastuzumab) | [130] | |
| Plasma | UC | RT-qPCR | mRNA | TrpC5, mdr1, MUC1 and flotillin2 | Predict chemoresistance (Anthracycline/taxan) | [135] | |
| Serum | Total Exosome Isolation Kit | RT-qPCR | mRNA | GSTP1 | Predict chemoresistance (Anthracycline/taxan) | [136] | |
| Ovarian cancer | Serum | UC + IP | RT-qPCR | miRNA | miR-21, miR-141, miR-200a, miR-200c, miR-200b, miR-203, miR-205, miR-214 | Diagnosis | [112] |
| Serum | Total Exosome Isolation Kit | RT-qPCR | miRNA | miR-1246 | Predict chemoresistance (Paclitaxel) | [218] | |
| Digestive cancer | |||||||
| Pancreatic cancer | Serum/Urine | UC | RT-qPCR | miRNA | miR-17-5p, miR-21 | Diagnosis | [219] |
| Plasma | ExoQuick | RT-qPCR | miRNA | miR-155 | Predict chemoresistance (Gemcitabine) | [131] | |
| Saliva | UC | RT-qPCR | mRNA | Apbb1ip, ASPN, Daf2, FoxP1, Bco31781, Gng2 | Diagnosis | [220] | |
| Liver cancer (HCC) | Plasma | Total Exosome Isolation Kit | RT-qPCR | tRNA | ValTAC-3, GlyTCC-5, ValAAC-5, GluCTC-5 | Diagnosis | [107] |
| Serum | Exoquick | RT-qPCR | circRNA | circUHRF1 | Diagnosis/immunotherapy resistance (anti-PD-1) | [108] | |
| Serum | Exoquick | RT-qPCR | miRNA | miR-21, miR-10b | Prognosis | [221] | |
| Gastoric cancer | Serum | unknown | RT-qPCR | miRNA | HOTTIP | Diagnosis/Prognosis | [124] |
| Serum | UC | RT-qPCR | lncRNA | lncUEGC | Diagnosis | [123] | |
| Serum | Exoquick | RT-qPCR | circRNA | circSHKBP1 | Diagnosis | [222] | |
| Esophageal cancer (ESCC) | Serum | Exoquick | RT-qPCR | miRNA | miR-21 | Diagnosis | [120] |
| Colorectal cancer | Serum | UC | RT-qPCR | miRNA | let-7a, miR-1229, miR-1246, miR-150, miR-21, miR-223, miR-23a | Diagnosis | [127] |
| Serum | ExoQuick | RT-qPCR | miRNA | miR-4772-3p | Prognosis | [223] | |
| Rectal cancer | Plasma | miRCURY Exosome Isolation Kit | RT-qPCR | miRNA | miR-30d-5p, miR-181a-5p and miR-486-5p | Diagnosis | [224] |
| Others | |||||||
| Lung cancer | Plasma | ExoQuick | RT-qPCR | miRNA | miR-151a-5p, miR-30a-3p, miR-200b-5p, miR-629, miR-100, and miR-154-3p | Diagnosis | [225] |
| Lung cancer (NSCLC) | Plasma | IP | RT-qPCR | miRNA | let-7f, miR-20b, miR-30e-3p, miR-223, miR-301 | Diagnosis | [226] |
| Lung cancer (LSCC) | Plasma | ExoQuick | RT-qPCR | miRNA | miR-205, miR-19a, miR-19b, miR-30b, miR-20a | Monitoring | [227] |
| Lung cancer (NSCLC) | Serum | UC | RT-qPCR | miRNA | miRNA-222-3p | Prognosis/Predict chemoresistance (Gemcitabine) | [228] |
| Lung cancer (NSCLC) | Serum | ExoQuick | RT-qPCR | miRNA | miRNA-146a-5p | Predict chemoresistance (Cisplatin) | [229] |
| Lung cancer (NSCLC) | Plasma | - | exoRNeasy kit/ddPCR | mRNA | PD-L1 | Predict immunotherapy resistance (anti-PD-1) | [134] |
| Melanoma | Plasma | - | exoRNeasy kit/ddPCR | mRNA | PD-L1 | Predict immunotherapy resistance (anti-PD-1) | [134] |
| Glioblastoma | Serum/Cereblospinal fluid | UC | RT-qPCR | miRNA | miR-21 | Diagnosis/Prognosis | [230] |
| Cereblospinal fluid | UC | RT-qPCR | miRNA | miR-21 | Diagnosis | [231] | |
| Serum | ExoQuick | RT-qPCR | miRNA | RNU6-1, miR-320, miR-574-3p | Diagnosis | [232] | |
| Serum | - | iMER | mRNA | MGMT, APNG | Predict chimoresistance (Temozolomide) | [137] |
EV RNAs for cancer diagnosis
Since the discovery of EV-miRNAs by Valadi et al.[55] in 2007, numerous studies have been performed to identify diagnostic EV miRNAs for cancer. Taylor et al.[112] showed that eight miRNAs (miR-21, miR-141, miR-200a, miR-200c, miR-200b, miR-203, miR-205 and miR-214) that were previously characterized as diagnostic markers for ovarian cancer were also upregulated in circulating EVs derived from ovarian cancer patients. Rabinowits et al.[113] conducted miRNA-profiling analyses of EV-based liquid biopsy samples and tumor biopsy samples from lung cancer patients and healthy controls. Their study showed a similarity in miRNA patterns between EV-based biopsy samples and tumor biopsy samples from lung cancer patients and demonstrated significant differences between these miRNA patterns and those in EVs from the healthy control group. This result indicates the potential of EV miRNAs as a liquid biopsy source for lung cancer. On the other hand, EV miR-1246 was significantly increased in ESCC patient serum samples but was not elevated in tumor biopsy samples, indicating that the level of EV miRNAs may not necessarily reflect the amounts in parental cells[114].
Regarding EVs in other body fluids, Nilsson et al.[115] reported that PCA3 and TMPRSS2:ERG mRNA, which are previously established prostate cancer biomarkers, were also present in urinary EVs from prostate cancer patients. Additionally, Foj et al.[116] demonstrated that the levels of miR-21-5p, miR-375, and let-7c-5p were remarkably increased in urinary EVs derived from prostate cancer patients compared with those from healthy subjects. Analyses of urinary EVs and associated RNAs are difficult to perform because of the low abundance of these molecules and the fact that the urine volume itself varies greatly over time. However, recently, a new technology has been reported in which zinc oxide nanowires are used to catch urinary EVs to increase the yield[117]. Moreover, ExoDx® Prostate (IntelliScore) is a simple, non-DRE, urine-based test for prostate cancer that is commercially available in the United States. This test has been clinically validated for risk stratification of clinically significant prostate cancer (Gleason score ≥ 7) from low-grade prostate cancer (Gleason score 6) and benign prostate disease, thus avoiding unnecessary prostate biopsy. A patient-specific individual risk score is evaluated based on an original algorithm that combines the expression level of three RNAs (PCA3 noncoding RNA, ERG mRNA, and SPDEF mRNA), which are correlated with clinically significant prostate cancer, detected directly in urinary EV RNAs[118].
To date, the potential of many miRNAs as cancer prognostic markers has been reported. Huang et al.[119] showed that increased levels of serum miR-1290 and miR-375 in EVs were correlated with decreased OS in patients with advanced-stage prostate cancer. Tanaka et al.[120] revealed that high levels of circulating EV miR-21 could distinguish esophageal squamous cell cancer (ESCC) patients from patients with benign diseases. The levels of EV miR-21 were also correlated with cancer progression and aggressiveness, indicating that EV miR-21 can serve as a diagnostic biomarker for cancer as well as a therapeutic target. In addition to EV miR-21, EV miR-1246 was also identified as a diagnostic and prognostic marker for ESCC[114]. Regarding prognostic markers for digestive cancers, Matsumura et al.[121]. reported that serum EV miR-19a-3p might be a prognostic biomarker to predict the recurrence of colorectal cancer. Other noncoding RNAs, such as lncRNAs and the less common family of circRNAs in EVs from cancer cells, have also attracted increasing attention. Urinary EV lncRNA p21 was reported to be elevated in prostate cancer patients and able to distinguish prostate cancer patients from those with benign disease[122]. Lin et al.[123] reported that lncRNA upregulated in plasma exosomes from gastric cancer (lncUEGC1) patients could be used to detect early-stage gastric cancer. Serum EV-associated lncRNA HOTTIP was also reported as a diagnostic and prognostic indicator of gastric cancer[124]. Furthermore, Lee et al.[125] demonstrated the prognostic significance of circulating EV ncRNAs [miRNA-21 and lncRNA activated by transforming growth factor beta (lncRNA-ATB)] for human hepatocellular carcinoma. In this study, the OS and PFS rates were significantly lower in patients with higher levels of miR-21 and lncRNA-ATB in EVs[125]. Concerning circRNA, Chen et al.[126] showed that circRNA PRMT5 was highly enriched in both serum and urinary EVs collected from patients with bladder cancer compared with normal individual cohorts. In this study, they also revealed that EV circPRMT5 levels were significantly correlated with cancer metastasis[126].
EV RNAs to monitor cancer progression and drug resistance
Similar to EV proteins, EV-RNAs have been reported to have potential as biomarkers for monitoring therapeutic effects or resistance to anticancer therapy.
Ogata-Kawata et al.[127] reported that the levels of seven miRNAs (let-7a, miR-1229, miR-1246, miR-150, miR-21, miR-223, miR-23a) in serum EVs were significantly increased in colorectal cancer patients. The serum levels of these miRNAs were significantly decreased after tumor resection, indicating their potential for monitoring tumor burden[127]. Svedman et al.[128] investigated EV-associated miRNA levels in patients with metastatic melanoma before, during, and after MAPK inhibitor therapy. They showed that increased levels of let-7g-5p were correlated with lower tumor burden and could clearly differentiate responders from nonresponders; moreover, an increase in the miR-497-5p level during the treatment course was significantly associated with better PFS rates[128]. Several researchers have investigated EV-associated lncRNAs in patients during chemotherapy. HOTAIR is one of the major lncRNAs that is overexpressed in a variety of cancers and promotes cancer cell proliferation, invasion and migration. Tang et al.[129] reported that EV HOTAIR was more enriched in serum samples from breast cancer patients than in healthy controls. In addition, they showed that a high pretreatment level of EV HOTAIR was correlated with a poor response to neoadjuvant chemotherapy and tamoxifen hormone therapy. In this study, EV HOTAIR was significantly decreased in all patients after surgery compared with before surgery, indicating that serum EV HOTAIR originates from the tumor tissue, and its level is associated with tumor burden and cancer aggressiveness[129].
Regarding EV lncRNAs, Dong et al.[130] have previously shown that isolated EVs from HER2-positive advanced breast cancer patients who were nonresponders to trastuzumab therapy contained more lncRNA SNHG14 than EVs from responders. In this study, the authors showed that EV lncRNA-SNHG14 promoted trastuzumab resistance by activating Bcl-2/apoptosis regulator BAX (Bax) signaling. Similar to this study, other types of EV RNAs have also been reported to be involved in resistance to anticancer therapy. Mikamori et al.[131] reported that higher miR-155 expression levels in resected tumor tissue samples from pancreatic ductal adenocarcinoma patients treated with gemcitabine (GEM) were correlated with a poorer prognosis. This study demonstrated that the levels of miR-155 in plasma-derived EVs were consistent with those in pancreatic tissue[131]. The results suggest that circulating EV biomarkers can reflect, to some extent, the tumor burden and possess potential as real-time monitoring biomarkers for anticancer drug resistance. Interestingly, they further demonstrated that high expression of miR-155 in pancreatic cancer cell lines promoted antiapoptotic signaling and EV secretion in vitro. Moreover, they showed that the EVs released by miR-155-overexpressing PDAC cell lines could transfer chemoresistance-associated molecules (including miR-155) to other cancer cells and that the recipient cells subsequently acquired chemoresistance to GEM in vitro. Del Re et al.[132] succeeded in detection of androgen receptor splice variant 7 (AR-V7) in EV RNAs from castration-resistant prostate cancer (CRPC) patients. The authors showed that EV AR-V7 mRNA levels were associated with hormonal therapy resistance. OS was significantly shorter among patients with EVs containing AR-V7 mRNA than among those without AR-V7 mRNA present (3 months vs. 20 months)[132]. Woo et al.[133] reported a practical method for analysis of AR-V7 mRNA in urinary EVs. In this study, the authors adopted a lab-on-a-disc integrated with six independent nanofiltration units, which enabled simultaneous processing of six individual samples. EV mRNA was extracted from each 4 mL urine sample, and AR-V7 and androgen receptor full-length (AR-FL) mRNA levels were quantified by dPCR. The results showed that higher AR-V7 and lower AR-FL expression was detected in urinary EVs derived from patients with CRPC than in those from patients with hormone-sensitive prostate cancer. In addition, they described that AR-V7 transcript levels and the AR-V7/AR-FL ratio in urinary EVs were higher in patients with advanced prostate cancer[133]. These studies indicate that EV AR-V7 mRNA could be a predictive biomarker for hormonal therapy resistance in prostate cancer. Some studies have investigated the relevance between EV RNAs and the response to ICIs. For example, Del Re et al[134]. demonstrated that EV-associated PD-L1 mRNA levels could be useful for real-time monitoring of the response to anti-PD-1 antibody therapy. They evaluated PD-L1 mRNA expression levels in plasma EVs from melanoma and NSCLC patients during nivolumab and pembrolizumab therapy. After 2 months of treatment, the EV PD-L1 mRNA copy numbers were significantly decreased in responders but remained unchanged in nonresponders[134]. Regarding the EV-based biomarkers associated with breast cancer chemoresistance, Ma et al.[135] performed profiling analyses of mRNA in circulating EVs from breast cancer patients treated with chemotherapy or not. The authors demonstrated that four mRNAs (TrpC5, MDR1, MUC1, and flotillin2) were amplified in EVs from patients with chemotherapy but were not amplified in patients without chemotherapy[135]. Interestingly, considering that TrpC5 was previously shown to regulate P-gp expression in recipient cells, this study suggested that TrpC5-enriched circulating EVs could promote cancer MDR development in these patients. More recently, Yang et al.[136] analyzed the levels of glutathione S-transferase P (GSTP1) mRNA in serum EVs from breast cancer patients treated with anthracycline/taxane-based neoadjuvant chemotherapy. GSTP1 is an enzyme that has a critical role in cell detoxification. Importantly, they observed that patients with EVs highly enriched in GSTP1 mRNA showed an inadequate response to chemotherapy[136].
Shao et al.[137] established a microfluidic platform termed immunomagnetic exosome RNA (iMER) analysis, which integrates immunomagnetic selection targeting EV-surface proteins and real-time qPCR for collecting EV-associated RNAs into a single microfluidic chip form. Serial measurements of the mRNA levels of MGMT and APNG, two important enzymes involved in repairing DNA damage induced by temozolomide for glioblastoma, demonstrated the feasibility of drug resistance monitoring during treatment using this integrated platform[137]. Although this kind of novel technology for capturing and analyzing EVs requires further development and validation in a clinical setting, it has the potential to reach the next level of EV utilization to detect cancer biomarkers.
EV DNA biomarkers
In addition to RNA species, DNA fragments have also been identified in EVs. Balaj et al.[138] were among the first to show that EVs contain single-stranded DNA. Yokoi et al.[139] confirmed the presence of double-stranded DNA in EVs using imaging flow cytometry and described how nuclear content was loaded into EVs. EV DNA is also a promising diagnostic tool due to its ability to carry information regarding cancer-specific mutations[140]. Here, we aim to summarize the growing evidence for EV DNA as a diagnostic marker and to consider its diagnostic advantages compared to cfDNA.
Kahlert et al.[141] identified the genomic DNA fragments in EVs from pancreatic cancer cell lines and pancreatic cancer patients. All genomic sequencing revealed mutations in KRAS and p53 in the genomic DNA of EVs derived from pancreatic cancer, suggesting that EV DNA sequencing can be used to determine treatment plans and predict therapy resistance[141]. However, in a subsequent paper, the first author called into question the superiority of EV DNA profiling to cfDNA profiling as an analyte for liquid biopsy[142]. Another remarkable study showed the detection rate of KRAS mutations in circulating EVs in PDAC patients and healthy controls. Mutations were detected in 7.4%, 66.7%, 80%, and 85% of age-matched controls and localized, locally advanced, and metastatic PDAC patients, respectively. In this study, the mutant KRAS detection rate in patients with localized PDAC after surgical resection dropped from 66.7% before surgery to 5% after surgery, indicating that EV-associated KRAS mutations could serve as biomarkers for real-time monitoring of therapy response and tumor burden[143]. A subsequent study demonstrated that the kinetics of EV-associated KRAS mutant allele frequency (MAF) were deeply correlated with neoadjuvant chemotherapy response; 71% of patients with a lack of cancer progression showed decreased KRAS MAF, while 94% of patients with cancer progression showed no decrease in KRAS MAF[144]. Interestingly, whereas the KRAS mutation detection rate in localized and metastatic pancreatic cancer was nearly equivalent for the cfDNA and EV DNA analyses, surgically resected primary tissue samples showed 95.5% concordance with the EV DNA-based assessment and only 68.2% concordance with the cfDNA analysis results. Similarly, agreement between the results for pancreatic patient tissue and liquid biopsy analyses was reported to be 83.3% for EV DNA-based analysis and only 66.8% for cfDNA-based analysis.
Similar to KRAS mutations, several analyses of circulating EVs from NSCLC patients showed the presence of clinically relevant epidermal growth factor receptor (EGFR)-specific mutations[145,146]. Remarkably, these studies highlight the expanded performance associated with combining analysis of EV-associated nucleic acids together with cfDNA vs. cfDNA analysis alone. Castellanos-Rizaldos et al.[145] reported that the combined use of EV nucleic acids together with cfDNA overcame the limited abundance of the EGFR T790M mutation and other EGFR mutations and contributed to improved sensitivity and specificity compared to cfDNA alone. Krug et al.[146] showed that the combined use of EV RNA and cfDNA sequencing improved the detection of EGFR mutations up to 98% vs. 84% for cfDNA alone. EV-based liquid biopsy has potential for multiplexing DNA analyses with analyses of other EV cargoes, such as miRNA, lncRNA, and proteins; thus, it can provide highly accurate information about cancer biology.
Other body fluids, such as urine, may also be valuable sources for EV DNA-based liquid biopsies. Lee et al.[147] investigated whether genetic alterations in urothelial bladder cancer were reflected in urinary cfDNA or EV DNA and demonstrated concordance between the copy number profiles of tumor tissue and urinary DNA (cfDNA and EV DNA), with allelic frequencies of 56.2% and 65.6%, respectively. Amplification of MDM2, ERBB2, CCND1, and CCNE1 and deletion of CDKN2A, PTEN, and RB1, whose alterations are all frequently found in bladder cancer, were also identified[147].
In addition to these studies, many studies have indicated that EV DNA may have a significant impact on the origin cells and recipient cells by playing a role in the maintenance of cellular homeostasis[148] and acting as an intercellular messenger[149]. Furthermore, similar to EV RNA, the packaging of DNA into membrane-enclosed vesicles contributes to enhanced stability by protecting it from the external environment and avoiding recognition by the immune system[150]. These findings may demonstrate that EV DNA is superior to cfDNA as a biomaterial in liquid biopsy for cancer; however, current protocols for definitively detecting cancer-derived EV DNA in clinical samples are hampered by high labor costs, high financial costs, and low accuracy. The ctDNA isolation and detection method has already been established and might be used in routine clinical situations in the near future[142]. Although cfDNA was first reported in the 1940s[151], the presence of EV DNA has long been doubted and was only demonstrated in the 2010s[138,140]. The potential of EV DNA as an analyte for liquid biopsies has not been thoroughly investigated but is a promising research area. We must improve strategies to utilize EV DNA and establish more useful methods of applying this molecule for liquid biopsy.
CHALLENGES AND FUTURE PERSPECTIVE
Most potential biomarkers are based on small-scale studies and require longitudinal validation in larger samples. In addition, the consistency and variability in data collected using different technologies are significant problems. The large amounts of data from recently developed detection technologies provide an opportunity to identify key molecules but also represent a challenge to differentiating valuable markers among numerous candidate molecules, and the associated processes need further investigation. Deeper and more rigorous studies are required to accurately correlate these potential markers with clinical practice. Moreover, the basic knowledge regarding the biological characteristics of EVs is still insufficient. The mechanisms that regulate the heterogeneity of cancer EVs have not been fully elucidated, and the factors that affect EV synthesis, secretion and transfer remain poorly understood; overcoming these issues is crucial to improving the accuracy of EV diagnostic outcomes.
The future of EV-based liquid biopsy depends on meeting certain technological issues. Isolation and detection of EVs are undoubtedly the biggest problems mentioned above; however, other technological problems have been observed. For example, RT-qPCR for EV-associated nucleic acids requires more appropriate housekeeping genes or reference genes, and universal genes that meet all the criteria as control genes for EV-associated nucleic acids have not yet been identified[152]. For EV-associated miRNAs, current protocols recommend that samples be processed from aligned volumes and that technical variations should be compensated for using synthetic nonhuman miRNAs, such as Caenorhabditis elegans cel-miR-39, as normalization controls. Variations can stem from many sources, such as differences in sample preparation, stabilization, RNA extraction, and target quantification. These differences are not a consequence of the disease state itself. Therefore, optimal genes that are stably expressed, irrespective of the experimental situation or treatment, must be identified to define reference genes for normalizing EV-associated nucleic acid expression. Several housekeeping genes or reference genes have been identified for different native tissues and body fluids, and stable endogenous RNAs have been proposed as internal controls; however, a consensus has not been reached[152]. In 2002, Vandesompele et al.[153] demonstrated that the common RT-qPCR procedure of using only one control gene induced relatively large errors. In this study, they claimed that ideal single internal control genes do not exist and recommended the use of at least three adequate control genes for calculating a normalization factor[153]. This issue with normalizers in gene expression analyses continues to this day, and the same issue is found with protein analyses. Quantification of EV-associated proteins also requires an internal control; however, the most suitable internal control for EV proteins for equivalent amounts of protein has not been identified. Not all EVs contain common EV marker proteins, such as Alix, TSG101, CD9, and CD63[154]. Hence, this problem with internal controls is a major problem associated with EV-based liquid biopsy, and further investigation is needed to develop biomarker research.
Despite these several concerns, EV-based liquid biopsy will provide higher sensitivity and specificity than classical biomarkers due to their stability in body fluids, and new technologies are being developed to solve the current limitations of EV-based liquid biopsy, as mentioned in this manuscript. Translating EV cargo profiles into routine clinical diagnostics would be facilitated by efficient alternatives to EV preparation via ultracentrifugation, such as biofluidic devices for high-throughput analysis. Moreover, extraordinary progress has occurred in analytic technologies, such as MS, HTS, and big-data analysis. These technologies have become irreplaceable and familiar analytical tools for researchers analyzing EV-associated molecules. Indeed, some studies have recommended MS-based methods as an alternative to detect EV protein markers after isolation procedures[155]. Further technological development will advance societal implementation of EV-based liquid biopsy for cancer.
CONCLUSIONS
Useful cancer biomarkers in liquid biopsies are urgently required, and EVs represent a promising resource for cancer biomarkers. The development of technologies is accompanied by novel statistical tools, which can utilize high-dimensional machine learning approaches to analyze big data and provide timely decisions. Thus, the future of EV-based liquid biopsy will be associated with multiple academic fields, such as molecular biology, bioengineering, clinical medicine, machine learning, and statistics. Numerous EV-associated studies will likely enhance the performance of cancer biomarkers in early diagnosis, prognosis, surveillance and treatment. Additionally, recent advances in bioinformatics may demonstrate the biological significance of identified cancer markers, which will provide clues for elucidation of cancer pathophysiology. In conclusion, the development of EV-based liquid biopsy will lead to early diagnosis of fatal cancers and tailor-made treatments for individual patients, and such advancements will extend the healthy human life span and reduce medical costs. Similar to the dramatic changes in our daily lives caused by a microscopic virus, a tiny vesicle may also dramatically advance cancer management.
DECLARATIONS
Authors’ contributionsDrafted the manuscript: Tamura T, Yoshioka Y, Ochiya T
Reviewed the manuscript: Yoshioka Y, Sakamoto S, Ichikawa T, Ochiya T
Approved the submitted manuscript: Yoshioka Y
All authors have read and agreed to the published version of the manuscript.
Availability of data and materialsNot applicable.
Financial support and sponsorshipThis work was supported by a Grant-in-Aid for Early Career Scientists from the Japan Society for the Promotion of Science (No. 19K16848, to Yoshioka Y), CREST from the Japan Science and Technology Agency (No. JPMJCR19H1, to Yoshioka Y) and Project for Cancer Research and Therapeutic Evolution (P-CREATE) from the Japan Agency for Medical Research and Development (No. JP20cm0106402, to Ochiya T).
Conflicts of interestThe authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2021.
REFERENCES
1. Doyle LM, Wang MZ. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019;8:727.
2. Siravegna G, Marsoni S, Siena S, Bardelli A. Integrating Liquid Biopsies into the Management of Cancer. Nat Rev Clin Oncol 2017;14:531-48.
5. Siravegna G, Mussolin B, Venesio T, et al. How liquid biopsies can change clinical practice in oncology. Ann Oncol 2019;30:1580-90.
6. Bettegowda C, Sausen M, Leary RJ, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med 2014;6:224ra24.
7. Cheng F, Su L, Qian C. Circulating tumor DNA: a promising biomarker in the liquid biopsy of cancer. Oncotarget 2016;7:48832-41.
8. Roy S, Hochberg FH, Jones PS. Extracellular vesicles: the growth as diagnostics and therapeutics; a Survey. J Extracell Vesicles 2018;7:1438720.
9. Yáñez-Mó M, Siljander PR, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 2015;4:27066.
10. Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 2015;25:981-4.
11. Keller S, Ridinger J, Rupp AK, Janssen JW, Altevogt P. Body fluid derived exosomes as a novel template for clinical diagnostics. J Transl Med 2011;9:86.
12. Krebs MG, Metcalf RL, Carter L, et al. Molecular analysis of circulating tumour cells - biology and biomarkers. Nat Rev Clin Oncol 2014;11:129-44.
13. Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol 2014;32:579-86.
14. Cristofanilli M, Budd GT, Ellis MJ, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004;351:781-91.
15. de Bono JS, Scher HI, Montgomery RB, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res 2008;14:6302-9.
16. Danila DC, Heller G, Gignac GA, et al. Circulating tumor cell number and prognosis in progressive castration-resistant prostate cancer. Clin Cancer Res 2007;13:7053-8.
17. Heller G, McCormack R, Kheoh T, et al. Circulating Tumor Cell Number as a Response Measure of Prolonged Survival for Metastatic Castration-Resistant Prostate Cancer: A Comparison with Prostate-Specific Antigen across Five Randomized Phase III Clinical Trials. J Clin Oncol 2018;36:572-80.
18. Goldkorn A, Ely B, Quinn DI, et al. Circulating tumor cell counts are prognostic of overall survival in SWOG S0421: a phase III trial of docetaxel with or without atrasentan for metastatic castration-resistant prostate cancer. J Clin Oncol 2014;32:1136-42.
19. Scher HI, Heller G, Molina A, et al. Circulating tumor cell biomarker panel as an individual-level surrogate for survival in metastatic castration-resistant prostate cancer. J Clin Oncol 2015;33:1348-55.
20. Allard WJ, Matera J, Miller MC, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res 2004;10:6897-904.
21. Alix-Panabières C, Pantel K. Circulating tumor cells: liquid biopsy of cancer. Clin Chem 2013;59:110-8.
22. Qin J, Alt JR, Hunsley BA, Williams TL, Fernando MR. Stabilization of circulating tumor cells in blood using a collection device with a preservative reagent. Cancer Cell Int 2014;14:23.
23. Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer 2014;14:623-31.
24. Allard WJ, Terstappen LWMM. CCR 20th anniversary commentary: paving the way for circulating tumor cells. Clin Cancer Res 2015;21:2883-5.
25. Markou A, Zavridou M, Sourvinou I, et al. Direct comparison of metastasis-related miRNAs expression levels in circulating tumor cells, corresponding plasma, and primary tumors of breast cancer patients. Clin Chem 2016;62:1002-11.
26. Antonarakis ES, Lu C, Luber B, et al. Clinical Significance of Androgen Receptor Splice Variant-7 mRNA Detection in Circulating Tumor Cells of Men With Metastatic Castration-Resistant Prostate Cancer Treated With First- and Second-Line Abiraterone and Enzalutamide. J Clin Oncol 2017;35:2149-56.
27. Sinkala E, Sollier-Christen E, Renier C, et al. Profiling protein expression in circulating tumour cells using microfluidic western blotting. Nat Commun 2017;8:14622.
28. Armbrecht L, Rutschmann O, Szczerba BM, Nikoloff J, Aceto N, Dittrich PS. Quantification of Protein Secretion from Circulating Tumor Cells in Microfluidic Chambers. Adv Sci (Weinh) 2020;7:1903237.
29. Zill OA, Banks KC, Fairclough SR, et al. The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients. Clin Cancer Res 2018;24:3528-38.
30. Chun FK, Müller I, Lange I, et al. Circulating tumour-associated plasma DNA represents an independent and informative predictor of prostate cancer. BJU Int 2006;98:544-8.
31. Altimari A, Grigioni AD, Benedettini E, et al. Diagnostic role of circulating free plasma DNA detection in patients with localized prostate cancer. Am J Clin Pathol 2008;129:756-62.
32. Bergsmedh A, Szeles A, Henriksson M, et al. Horizontal transfer of oncogenes by uptake of apoptotic bodies. Proc Natl Acad Sci U S A 2001;98:6407-11.
33. Trejo-Becerril C, Pérez-Cárdenas E, Taja-Chayeb L, et al. Cancer progression mediated by horizontal gene transfer in an in vivo model. PLoS One 2012;7:e52754.
34. Mahon KL, Qu W, Devaney J, et al. PRIMe consortium. Methylated Glutathione S-transferase 1 (mGSTP1) is a potential plasma free DNA epigenetic marker of prognosis and response to chemotherapy in castrate-resistant prostate cancer. Br J Cancer 2014;111:1802-9.
35. Azad AA, Volik SV, Wyatt AW, et al. Androgen Receptor Gene Aberrations in Circulating Cell-Free DNA: Biomarkers of Therapeutic Resistance in Castration-Resistant Prostate Cancer. Clin Cancer Res 2015;21:2315-24.
36. Lallous N, Volik SV, Awrey S, et al. Functional analysis of androgen receptor mutations that confer anti-androgen resistance identified in circulating cell-free DNA from prostate cancer patients. Genome Biol 2016;17:10.
37. Salvi S, Casadio V, Conteduca V, et al. Circulating cell-free AR and CYP17A1 copy number variations may associate with outcome of metastatic castration-resistant prostate cancer patients treated with abiraterone. Br J Cancer 2015;112:1717-24.
38. Carreira S, Romanel A, Goodall J, et al. Tumor clone dynamics in lethal prostate cancer. Sci Transl Med 2014;6:254ra125.
39. Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med 2008;14:985-90.
40. Leung F, Kulasingam V, Diamandis EP, et al. Circulating tumor DNA as a cancer biomarker: fact or fiction? Clin Chem 2016;62:1054-60.
41. Sozzi G, Roz L, Conte D, et al. Effects of prolonged storage of whole plasma or isolated plasma DNA on the results of circulating DNA quantification assays. J Natl Cancer Inst 2005;97:1848-50.
42. Gormally E, Caboux E, Vineis P, Hainaut P. Circulating free DNA in plasma or serum as biomarker of carcinogenesis: practical aspects and biological significance. Mutat Res 2007;635:105-17.
43. Cai X, Janku F, Zhan Q, Fan JB. Accessing genetic information with liquid biopsies. Trends Genet 2015;31:564-75.
44. Schwarzenbach H, Hoon DSB, Pantel K. Cell-free nucleic acids as biomarkers in cancer patients. Nat Rev Cancer 2011;11:426-37.
45. Gould SJ, Raposo G. As we wait: coping with an imperfect nomenclature for extracellular vesicles. J Extracell Vesicles 2013;2:20389.
46. György B, Szabó TG, Pásztói M, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci 2011;68:2667-88.
47. Théry C, Witwer KW, Aikawa E, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018;7:1535750.
49. György B, Szabó TG, Pásztói M, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci 2011;68:2667-88.
50. Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013;200:373-83.
51. Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci U S A 2016;113:E968-77.
52. Pan B, Johnstone RM. Fate of the transferrin receptor during maturation of sheep reticulocytes in vitro: Selective externalization of the receptor. Cell 1983;33:967-78.
53. Johnstone RM, Adam M, Hammond JR, Orr L, Turbide C. Vesicle formation during reticulocyte maturation. association of plasma membrane activities with released vesicles (exosomes). J Biol Chem 1987:262;9412-20.
54. Raposo G, Nijman HW, Stoorvogel W, et al. B lymphocytes secrete antigen-presenting vesicles. J Exp Med 1996;183:1161-72.
55. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9.
56. Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature 2016;537:347-55.
57. Reuter JA, Spacek DV, Snyder MP. High-throughput sequencing technologies. Mol Cell 2015;58:586-97.
58. Welch JL, Madison MN, Margolick JB, et al. Effect of prolonged freezing of semen on exosome recovery and biologic activity. Sci Rep 2017;7:45034.
59. Tovar-Camargo OA, Toden S, Goel A. Exosomal microRNA biomarkers: emerging frontiers in colorectal and other human cancers. Expert Rev Mol Diagn 2016;16:553-67.
60. Li P, Kaslan M, Lee SH, Yao J, Gao Z. Progress in exosome isolation techniques. Theranostics 2017;7:789-804.
61. Nordin JZ, Lee Y, Vader P, et al. Ultrafiltration with size-exclusion liquid chromatography for high yield isolation of extracellular vesicles preserving intact biophysical and functional properties. Nanomedicine 2015;11:879-83.
62. Gupta S, Rawat S, Arora V, et al. An improvised one-step sucrose cushion ultracentrifugation method for exosome isolation from culture supernatants of mesenchymal stem cells. Stem Cell Res Ther 2018;9:180.
63. Van Deun J, Mestdagh P, Sormunen R, et al. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J Extracell Vesicles 2014;3:24858.
64. Coumans FAW, Brisson AR, Buzas EI, et al. Methodological guidelines to study extracellular vesicles. Circ Res 2017;120:1632-48.
65. Helwa I, Cai J, Drewry MD, et al. A Comparative Study of Serum Exosome Isolation Using Differential Ultracentrifugation and Three Commercial Reagents. PLoS One 2017;12:e0170628.
66. Clayton A, Court J, Navabi H, et al. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J Immunol Methods 2001;247:163-74.
67. Théry C, Amigorena S, Raposo G, Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol 2006;Chapter 3:Unit 3.22.
68. Brennan K, Martin K, FitzGerald SP, et al. A comparison of methods for the isolation and separation of extracellular vesicles from protein and lipid particles in human serum. Sci Rep 2020;10:1039.
69. Liangsupree T, Multia E, Riekkola ML. Modern isolation and separation techniques for extracellular vesicles. J Chromatogr A 2021;1636:461773.
70. An M, Wu J, Zhu J, Lubman DM. Comparison of an Optimized Ultracentrifugation Method versus Size-Exclusion Chromatography for Isolation of Exosomes from Human Serum. J Proteome Res 2018;17:3599-605.
71. Koh YQ, Almughlliq FB, Vaswani K, Peiris HN, Mitchell MD. Exosome enrichment by ultracentrifugation and size exclusion chromatography. Front Biosci (Landmark Ed) 2018;23:865-74.
72. Wei R, Zhao L, Kong G, et al. Combination of Size-Exclusion Chromatography and Ultracentrifugation Improves the Proteomic Profiling of Plasma-Derived Small Extracellular Vesicles. Biol Proced Online 2020;22:12.
73. Simons M, Raposo G. Exosomes - vesicular carriers for intercellular communication. Curr Opin Cell Biol 2009;21:575-81.
74. Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics 2010;73:1907-20.
75. Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol 2014;29:116-25.
76. Clayton A, Boilard E, Buzas EI, et al. Considerations towards a roadmap for collection, handling and storage of blood extracellular vesicles. J Extracell Vesicles 2019;8:1647027.
77. Jørgensen M, Bæk R, Pedersen S, Søndergaard EK, Kristensen SR, Varming K. Extracellular Vesicle (EV) Array: microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping. J Extracell Vesicles 2013;2:20920.
78. Jakobsen KR, Paulsen BS, Bæk R, Varming K, Sorensen BS, Jørgensen MM. Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma. J Extracell Vesicles 2015;4:26659.
79. Shao H, Chung J, Balaj L, et al. Protein typing of circulating microvesicles allows real-time monitoring of glioblastoma therapy. Nat Med 2012;18:1835-40.
80. Kravets VG, Schedin F, Jalil R, et al. Singular phase nano-optics in plasmonic metamaterials for label-free single-molecule detection. Nat Mater 2013;12:304-9.
81. Im H, Shao H, Park YI, et al. Label-free detection and molecular profiling of exosomes with a nano-plasmonic sensor. Nat Biotechnol 2014;32:490-5.
82. Yoshioka Y, Kosaka N, Konishi Y, et al. Ultra-sensitive liquid biopsy of circulating extracellular vesicles using ExoScreen. Nat Commun 2014;5:3591.
83. Zhao Z, Yang Y, Zeng Y, He M. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip 2016;16:489-96.
84. Mitchell PJ, Welton J, Staffurth J, et al. Can urinary exosomes act as treatment response markers in prostate cancer? J Transl Med 2009;7:4.
85. Logozzi M, Angelini DF, Iessi E, et al. Increased PSA expression on prostate cancer exosomes in in vitro condition and in cancer patients. Cancer Lett 2017;403:318-29.
86. Huber V, Fais S, Iero M, et al. Human colorectal cancer cells induce T-cell death through release of proapoptotic microvesicles: role in immune escape. Gastroenterology 2005;128:1796-804.
87. Khan S, Jutzy JM, Valenzuela MM, et al. Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS One 2012;7:e46737.
88. Khan S, Bennit HF, Turay D, et al. Early diagnostic value of survivin and its alternative splice variants in breast cancer. BMC Cancer 2014;14:176.
89. Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015;523:177-82.
91. Giampieri R, Piva F, Occhipinti G, et al. Clinical impact of different exosomes' protein expression in pancreatic ductal carcinoma patients treated with standard first line palliative chemotherapy. PLoS One 2019;14:e0215990.
92. Ning K, Wang T, Sun X, et al. UCH-L1-containing exosomes mediate chemotherapeutic resistance transfer in breast cancer. J Surg Oncol 2017;115:932-40.
93. Lv MM, Zhu XY, Chen WX, et al. Exosomes mediate drug resistance transfer in MCF-7 breast cancer cells and a probable mechanism is delivery of P-glycoprotein. Tumour Biol 2014;35:10773-9.
94. Zhang FF, Zhu YF, Zhao QN, et al. Microvesicles mediate transfer of P-glycoprotein to paclitaxel-sensitive A2780 human ovarian cancer cells, conferring paclitaxel-resistance. Eur J Pharmacol 2014;738:83-90.
95. Corcoran C, Rani S, O'Brien K, et al. Docetaxel-resistance in prostate cancer: evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS One 2012;7:e50999.
96. Ciravolo V, Huber V, Ghedini GC, et al. Potential role of HER2-overexpressing exosomes in countering trastuzumab-based therapy. J Cell Physiol 2012;227:658-67.
97. Martinez VG, O'Neill S, Salimu J, et al. Resistance to HER2-targeted anti-cancer drugs is associated with immune evasion in cancer cells and their derived extracellular vesicles. Oncoimmunology 2017;6:e1362530.
98. Chen L, Han X. Anti-PD-1/PD-L1 therapy of human cancer: past, present, and future. J Clin Invest 2015;125:3384-91.
99. Ribas A, Hamid O, Daud A, et al. Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA 2016;315:1600-9.
100. Zaretsky JM, Garcia-Diaz A, Shin DS, et al. Mutations Associated with Acquired Resistance to PD-1 Blockade in Melanoma. N Engl J Med 2016;375:819-29.
101. Chen G, Huang AC, Zhang W, et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018;560:382-6.
102. Cao L, Cui X, Hu J, et al. Advances in digital polymerase chain reaction (dPCR) and its emerging biomedical applications. Biosens Bioelectron 2017;90:459-74.
103. Ratajczak J, Miekus K, Kucia M, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006;20:847-56.
104. Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem 2010;285:17442-52.
105. Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A 2010;107:6328-33.
106. Zhang Y, Liu D, Chen X, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell 2010;39:133-44.
107. Zhu L, Li J, Gong Y, et al. Exosomal tRNA-derived small RNA as a promising biomarker for cancer diagnosis. Mol Cancer 2019;18:74.
108. Zhang PF, Gao C, Huang XY, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer 2020;19:110.
109. Gusachenko ON, Zenkova MA, Vlassov VV. Nucleic acids in exosomes: disease markers and intercellular communication molecules. Biochem 2013;78:1-7.
110. Bullock MD, Silva AM, Kanlikilicer-Unaldi P, et al. Exosomal non-coding RNAs: diagnostic, prognostic and therapeutic applications in cancer. Noncoding RNA 2015;1:53-68.
111. Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 2008;105:10513-8.
112. Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol 2008;110:13-21.
113. Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer 2009;10:42-6.
114. Takeshita N, Hoshino I, Mori M, et al. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer 2013;108:644-52.
115. Nilsson J, Skog J, Nordstrand A, et al. Prostate cancer-derived urine exosomes: a novel approach to biomarkers for prostate cancer. Br J Cancer 2009;100:1603-7.
116. Foj L, Ferrer F, Serra M, et al. Exosomal and Non-Exosomal Urinary miRNAs in Prostate Cancer Detection and Prognosis. Prostate 2017;77:573-83.
117. Yasui T, Yanagida T, Ito S, et al. Unveiling massive numbers of cancer-related urinary-microRNA candidates via nanowires. Sci Adv 2017;3:e1701133.
118. McKiernan J, Donovan MJ, O'Neill V, et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol 2016;2:882-9.
119. Huang X, Yuan T, Liang M, et al. Exosomal miR-1290 and miR-375 as prognostic markers in castration-resistant prostate cancer. Eur Urol 2015;67:33-41.
120. Tanaka Y, Kamohara H, Kinoshita K, et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer 2013;119:1159-67.
121. Matsumura T, Sugimachi K, Iinuma H, et al. Exosomal microRNA in serum is a novel biomarker of recurrence in human colorectal cancer. Br J Cancer 2015;113:275-81.
122. Işın M, Uysaler E, Özgür E, et al. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front Genet 2015;6:168.
123. Lin LY, Yang L, Zeng Q, et al. Tumor-originated exosomal LncUEGC1 as a circulating biomarker for early-stage gastric cancer. Mol Cancer 2018;17:84.
124. Zhao R, Zhang Y, Zhang X, et al. Exosomal long noncoding RNA HOTTIP as potential novel diagnostic and prognostic biomarker test for gastric cancer. Mol Cancer 2018;17:68.
125. Lee YR, Kim G, Tak WY, et al. Circulating exosomal noncoding RNAs as prognostic biomarkers in human hepatocellular carcinoma. Int J Cancer 2019;144:1444-52.
126. Chen X, Chen RX, Wei WS, et al. PRMT5 Circular RNA Promotes Metastasis of Urothelial Carcinoma of the Bladder through Sponging miR-30c to Induce Epithelial-Mesenchymal Transition. Clin Cancer Res 2018;24:6319-30.
127. Ogata-Kawata H, Izumiya M, Kurioka D, et al. Circulating exosomal microRNAs as biomarkers of colon cancer. PLoS One 2014;9:e92921.
128. Svedman FC, Lohcharoenkal W, Bottai M, et al. Extracellular microvesicle microRNAs as predictive biomarkers for targeted therapy in metastastic cutaneous malignant melanoma. PLoS One 2018;13:e0206942.
129. Tang S, Zheng K, Tang Y, Li Z, Zou T, Liu D. Overexpression of serum exosomal HOTAIR is correlated with poor survival and poor response to chemotherapy in breast cancer patients. J Biosci 2019:44.
130. Dong H, Wang W, Chen R, et al. Exosome-mediated transfer of lncRNASNHG14 promotes trastuzumab chemoresistance in breast cancer. Int J Oncol 2018;53:1013-26.
131. Mikamori M, Yamada D, Eguchi H, et al. MicroRNA-155 Controls Exosome Synthesis and Promotes Gemcitabine Resistance in Pancreatic Ductal Adenocarcinoma. Sci Rep 2017;7:42339.
132. Del Re M, Biasco E, Crucitta S, et al. The Detection of Androgen Receptor Splice Variant 7 in Plasma-derived Exosomal RNA Strongly Predicts Resistance to Hormonal Therapy in Metastatic Prostate Cancer Patients. Eur Urol 2017;71:680-7.
133. Woo HK, Park J, Ku JY, et al. Urine-based liquid biopsy: non-invasive and sensitive AR-V7 detection in urinary EVs from patients with prostate cancer. Lab Chip 2018;19:87-97.
134. Del Re M, Marconcini R, Pasquini G, et al. PD-L1 mRNA expression in plasma-derived exosomes is associated with response to anti-PD-1 antibodies in melanoma and NSCLC. Br J Cancer 2018;118:820-4.
135. Ma X, Chen Z, Hua D, et al. Essential role for TrpC5-containing extracellular vesicles in breast cancer with chemotherapeutic resistance. Proc Natl Acad Sci U S A 2014;111:6389-94.
136. Yang SJ, Wang DD, Li J, et al. Predictive role of GSTP1-containing exosomes in chemotherapy-resistant breast cancer. Gene 2017;623:5-14.
137. Shao H, Chung J, Lee K, et al. Chip-based analysis of exosomal mRNA mediating drug resistance in glioblastoma. Nat Commun 2015;6:6999.
138. Balaj L, Lessard R, Dai L, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun 2011;2:180.
139. Yokoi A, Villar-Prados A, Oliphint PA, et al. Mechanisms of nuclear content loading to exosomes. Sci Adv 2019;5:eaax8849.
140. Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 2014;24:766-9.
141. Kahlert C, Melo SA, Protopopov A, et al. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J Biol Chem 2014;289:3869-75.
142. Kahlert C. Liquid Biopsy: Is There an Advantage to Analyzing Circulating Exosomal DNA Compared to cfDNA or Are They the Same? Cancer Res 2019;79:2462-5.
143. Allenson K, Castillo J, San Lucas FA, et al. High prevalence of mutant KRAS in circulating exosome-derived DNA from early-stage pancreatic cancer patients. Ann Oncol 2017;28:741-7.
144. Bernard V, Kim DU, San Lucas FA, et al. Circulating Nucleic Acids Are Associated With Outcomes of Patients With Pancreatic Cancer. Gastroenterology 2019;156:108-118.e4.
145. Castellanos-Rizaldos E, Grimm DG, Tadigotla V, et al. Exosome-Based Detection of. EGFR ;24:2944-50.
146. Krug AK, Enderle D, Karlovich C, et al. Improved EGFR mutation detection using combined exosomal RNA and circulating tumor DNA in NSCLC patient plasma. Ann Oncol 2018;29:2143.
147. Lee DH, Yoon H, Park S, et al. Urinary Exosomal and cell-free DNA Detects Somatic Mutation and Copy Number Alteration in Urothelial Carcinoma of Bladder. Sci Rep 2018;8:14707.
148. Takahashi A, Okada R, Nagao K, et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat Commun 2017;8:15287.
149. Fischer S, Cornils K, Speiseder T, et al. Indication of Horizontal DNA Gene Transfer by Extracellular Vesicles. PLoS One 2016;11:e0163665.
150. Jin Y, Chen K, Wang Z, et al. DNA in serum extracellular vesicles is stable under different storage conditions. BMC Cancer 2016;16:753.
151. Mandel P, Métais P. Les Acides Nucleiques Du Plasma Sanguin Chez l’ Homme. C R Seances Soc Biol Fil 1948;142:241-3. (in French).
152. Schwarzenbach H, da Silva AM, Calin G, Pantel K. Data Normalization Strategies for MicroRNA Quantification. Clin Chem 2015;61:1333-42.
153. Vandesompele J, De Preter K, Pattyn F, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol 2002;3:RESEARCH0034.
154. Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics 2009;9:4997-5000.
155. Nguyen HQ, Lee D, Kim Y, et al. Platelet Factor 4 as a Novel Exosome Marker in MALDI-MS Analysis of Exosomes from Human Serum. Anal Chem 2019;91:13297-305.
156. Lu Q, Zhang J, Allison R, et al. Identification of extracellular delta-catenin accumulation for prostate cancer detection. Prostate 2009;69:411-8.
157. Kharaziha P, Chioureas D, Rutishauser D, et al. Molecular profiling of prostate cancer derived exosomes may reveal a predictive signature for response to docetaxel. Oncotarget 2015;6:21740-54.
158. Biggs CN, Siddiqui KM, Al-Zahrani AA, et al. Prostate extracellular vesicles in patient plasma as a liquid biopsy platform for prostate cancer using nanoscale flow cytometry. Oncotarget 2016;7:8839-49.
159. Smalley DM, Sheman NE, Nelson K, Theodorescu D. Isolation and identification of potential urinary microparticle biomarkers of bladder cancer. J Proteome Res 2008;7:2088-96.
160. Chen CL, Lai YF, Tang P, et al. Comparative and targeted proteomic analyses of urinary microparticles from bladder cancer and hernia patients. J Proteome Res 2012;11:5611-29.
161. Lin SY, Chang CH, Wu HC, et al. Proteome Profiling of Urinary Exosomes Identifies Alpha 1-Antitrypsin and H2B1K as Diagnostic and Prognostic Biomarkers for Urothelial Carcinoma. Sci Rep 2016;6:34446.
162. Silvers CR, Miyamoto H, Messing EM, Netto GJ, Lee YF. Characterization of urinary extracellular vesicle proteins in muscle-invasive bladder cancer. Oncotarget 2017;8:91199-208.
163. Silvers CR, Liu YR, Wu CH, Miyamoto H, Messing EM, Lee YF. Identification of extracellular vesicle-borne periostin as a feature of muscle-invasive bladder cancer. Oncotarget 2016;7:23335-45.
164. Beckham CJ, Olsen J, Yin PN, et al. Bladder cancer exosomes contain EDIL-3/Del1 and facilitate cancer progression. J Urol 2014;192:583-92.
165. Welton JL, Khanna S, Giles PJ, et al. Proteomics analysis of bladder cancer exosomes. Mol Cell Proteomics 2010;9:1324-38.
166. Raimondo F, Morosi L, Corbetta S, et al. Differential protein profiling of renal cell carcinoma urinary exosomes. Mol Biosyst 2013;9:1220-33.
167. Wang X, Zhong W, Bu J, et al. Exosomal protein CD82 as a diagnostic biomarker for precision medicine for breast cancer. Mol Carcinog 2019;58:674-85.
168. Rupp AK, Rupp C, Keller S, et al. Loss of EpCAM expression in breast cancer derived serum exosomes: role of proteolytic cleavage. Gynecol Oncol 2011;122:437-46.
169. Wang T, Ning K, Lu TX, et al. Increasing circulating exosomes-carrying TRPC5 predicts chemoresistance in metastatic breast cancer patients. Cancer Sci 2017;108:448-54.
170. Szajnik M, Derbis M, Lach M, et al. Exosomes in Plasma of Patients with Ovarian Carcinoma: Potential Biomarkers of Tumor Progression and Response to Therapy. Gynecol Obstet (Sunnyvale) 2013;Suppl 4:3.
171. Li J, Sherman-Baust CA, Tsai-Turton M, Bristow RE, Roden RB, Morin PJ. Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer. BMC Cancer 2009;9:244.
172. Tang MKS, Yue PYK, Ip PP, et al. Soluble E-cadherin promotes tumor angiogenesis and localizes to exosome surface. Nat Commun 2018;9:2270.
173. Graves LE, Ariztia EV, Navari JR, Matzel HJ, Stack MS, Fishman DA. Proinvasive properties of ovarian cancer ascites-derived membrane vesicles. Cancer Res 2004;64:7045-9.
174. Keller S, König AK, Marmé F, et al. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Lett 2009;278:73-81.
175. Frampton AE, Prado MM, López-Jiménez E, et al. Glypican-1 is enriched in circulating-exosomes in pancreatic cancer and correlates with tumor burden. Oncotarget 2018;9:19006-13.
176. Kimura H, Yamamoto H, Harada T, et al. CKAP4, a DKK1 Receptor, Is a Biomarker in Exosomes Derived from Pancreatic Cancer and a Molecular Target for Therapy. Clin Cancer Res 2019;25:1936-47.
177. Costa-Silva B, Aiello NM, Ocean AJ, et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat Cell Biol 2015;17:816-26.
178. Yokoyama S, Takeuchi A, Yamaguchi S, et al. Clinical implications of carcinoembryonic antigen distribution in serum exosomal fraction-Measurement by ELISA. PLoS One 2017;12:e0183337.
179. Choi DS, Park JO, Jang SC, et al. Proteomic analysis of microvesicles derived from human colorectal cancer ascites. Proteomics 2011;11:2745-51.
180. Baran J, Baj-Krzyworzeka M, Weglarczyk K, et al. Circulating tumour-derived microvesicles in plasma of gastric cancer patients. Cancer Immunol Immunother 2010;59:841-50.
181. Yamashita T, Kamada H, Kanasaki S, et al. Epidermal growth factor receptor localized to exosome membranes as a possible biomarker for lung cancer diagnosis. Pharmazie 2013:68;969-73.
182. Niu L, Song X, Wang N, Xue L, Song X, Xie L. Tumor-derived exosomal proteins as diagnostic biomarkers in non-small cell lung cancer. Cancer Sci 2019;110:433-42.
183. Sandfeld-Paulsen B, Aggerholm-Pedersen N, Bæk R, et al. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol Oncol 2016;10:1595-602.
184. Li Y, Zhang Y, Qiu F, Qiu Z. Proteomic identification of exosomal LRG1: a potential urinary biomarker for detecting NSCLC. Electrophoresis 2011;32:1976-83.
185. Logozzi M, De Milito A, Lugini L, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One 2009;4:e5219.
186. Peinado H, Alečković M, Lavotshkin S, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med 2012;18:883-91.
187. Bryant RJ, Pawlowski T, Catto JW, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer 2012;106:768-74.
188. Xu Y, Qin S, An T, Tang Y, Huang Y, Zheng L. MiR-145 detection in urinary extracellular vesicles increase diagnostic efficiency of prostate cancer based on hydrostatic filtration dialysis method. Prostate 2017;77:1167-75.
189. Koppers-Lalic D, Hackenberg M, de Menezes R, et al. Noninvasive prostate cancer detection by measuring miRNA variants (isomiRs) in urine extracellular vesicles. Oncotarget 2016;7:22566-78.
190. Wani S, Kaul D, Mavuduru RS, Kakkar N, Bhatia A. Urinary-exosomal miR-2909: A novel pathognomonic trait of prostate cancer severity. J Biotechnol 2017;259:135-9.
191. Rodríguez M, Bajo-Santos C, Hessvik NP, et al. Identification of non-invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol Cancer 2017;16:1-6.
192. Endzeliņš E, Berger A, Melne V, et al. Detection of circulating miRNAs: comparative analysis of extracellular vesicle-incorporated miRNAs and cell-free miRNAs in whole plasma of prostate cancer patients. BMC Cancer 2017;17:730.
193. Li Z, Ma YY, Wang J, et al. Exosomal microRNA-141 is upregulated in the serum of prostate cancer patients. Onco Targets Ther 2016;9:139-48.
194. Hessvik NP, Sandvig K, Llorente A. Exosomal miRNAs as Biomarkers for Prostate Cancer. Front Genet 2013;4:36.
195. Moltzahn F, Olshen AB, Baehner L, et al. Microfluidic-based multiplex qRT-PCR identifies diagnostic and prognostic microRNA signatures in the sera of prostate cancer patients. Cancer Res 2011;71:550-60.
196. Lodes MJ, Caraballo M, Suciu D, Munro S, Kumar A, Anderson B. Detection of cancer with serum miRNAs on an oligonucleotide microarray. PLoS One 2009;4:e6229.
197. Bhagirath D, Yang TL, Bucay N, et al. microRNA-1246 Is an Exosomal Biomarker for Aggressive Prostate Cancer. Cancer Res 2018;78:1833-44.
198. Wang YH, Ji J, Wang BC, et al. Tumor-Derived Exosomal Long Noncoding RNAs as Promising Diagnostic Biomarkers for Prostate Cancer. Cell Physiol Biochem 2018;46:532-45.
199. De Long J, Sullivan TB, Humphrey J, et al. A non-invasive miRNA based assay to detect bladder cancer in cell-free urine. Am J Transl Res 2015;7:2500-9.
200. Armstrong DA, Green BB, Seigne JD, Schned AR, Marsit CJ. MicroRNA molecular profiling from matched tumor and bio-fluids in bladder cancer. Mol Cancer 2015;14:194.
201. Andreu Z, Otta Oshiro R, Redruello A, et al. Extracellular vesicles as a source for non-invasive biomarkers in bladder cancer progression. Eur J Pharm Sci 2017;98:70-9.
202. Baumgart S, Meschkat P, Edelmann P, et al. MP78-05 INVASION-ASSOCIATED MIRNAS S AS POSSIBLE DIAGNOSTIC BIOMARKERS OF MUSCLE INVASIVE BLADDER CANCER IN TUMOR TISSUES AND URINARY EXOSOMES. J Urol 2018:199.
203. Berrondo C, Flax J, Kucherov V, et al. Expression of the Long Non-Coding RNA HOTAIR Correlates with Disease Progression in Bladder Cancer and Is Contained in Bladder Cancer Patient Urinary Exosomes. PLoS One 2016;11:e0147236.
204. Zhang S, Du L, Wang L, et al. Evaluation of serum exosomal LncRNA-based biomarker panel for diagnosis and recurrence prediction of bladder cancer. J Cell Mol Med 2019;23:1396-405.
205. Xue M, Chen W, Xiang A, et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol Cancer 2017;16:143.
206. Zhang W, Ni M, Su Y, et al. MicroRNAs in Serum Exosomes as Potential Biomarkers in Clear-cell Renal Cell Carcinoma. Eur Urol Focus 2018;4:412-9.
207. Wang X, Wang T, Chen C, et al. Serum exosomal miR-210 as a potential biomarker for clear cell renal cell carcinoma. J Cell Biochem ;2018:1492-502.
208. Butz H, Nofech-Mozes R, Ding Q, et al. Exosomal MicroRNAs Are Diagnostic Biomarkers and Can Mediate Cell-Cell Communication in Renal Cell Carcinoma. Eur Urol Focus 2016;2:210-8.
209. Fujii N, Hirata H, Ueno K, et al. Extracellular MiR-224 as a prognostic marker for clear cell renal cell carcinoma. Oncotarget 2017;8:109877-88.
210. Du M, Giridhar KV, Tian Y, et al. Plasma exosomal miRNAs-based prognosis in metastatic kidney cancer. Oncotarget 2017;8:63703-14.
211. De Palma G, Sallustio F, Curci C, et al. The Three-Gene Signature in Urinary Extracellular Vesicles from Patients with Clear Cell Renal Cell Carcinoma. J Cancer 2016;7:1960-7.
212. Hannafon BN, Trigoso YD, Calloway CL, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res 2016;18:90.
213. Zhou W, Fong MY, Min Y, et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014;25:501-15.
214. Rodríguez-Martínez A, de Miguel-Pérez D, Ortega FG, et al. Exosomal miRNA profile as complementary tool in the diagnostic and prediction of treatment response in localized breast cancer under neoadjuvant chemotherapy. Breast Cancer Res 2019;21:21.
215. Santos JC, Lima NDS, Sarian LO, Matheu A, Ribeiro ML, Derchain SFM. Exosome-mediated breast cancer chemoresistance via miR-155 transfer. Sci Rep 2018;8:829.
216. Gregory PA, Bert AG, Paterson EL, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008;10:593-601.
217. Pigati L, Yaddanapudi SC, Iyengar R, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One 2010;5:e13515.
218. Kanlikilicer P, Bayraktar R, Denizli M, et al. Exosomal miRNA confers chemo resistance via targeting Cav1/p-gp/M2-type macrophage axis in ovarian cancer. EBioMedicine 2018;38:100-12.
219. Que R, Ding G, Chen J, Cao L. Analysis of serum exosomal microRNAs and clinicopathologic features of patients with pancreatic adenocarcinoma. World J Surg Oncol 2013;11:219.
220. Lau C, Kim Y, Chia D, et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J Biol Chem 2013;288:26888-97.
221. Tian XP, Wang CY, Jin XH, et al. Acidic Microenvironment Up-Regulates Exosomal miR-21 and miR-10b in Early-Stage Hepatocellular Carcinoma to Promote Cancer Cell Proliferation and Metastasis. Theranostics 2019;9:1965-79.
222. Xie M, Yu T, Jing X, et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer 2020;19:112.
223. Liu C, Eng C, Shen J, et al. Serum exosomal miR-4772-3p is a predictor of tumor recurrence in stage II and III colon cancer. Oncotarget 2016;7:76250-60.
224. Bjørnetrø T, Redalen KR, Meltzer S, et al. An experimental strategy unveiling exosomal microRNAs 486-5p, 181a-5p and 30d-5p from hypoxic tumour cells as circulating indicators of high-risk rectal cancer. J Extracell Vesicles 2019;8:1567219.
225. Cazzoli R, Buttitta F, Di Nicola M, et al. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J Thorac Oncol 2013;8:1156-62.
226. Silva J, García V, Zaballos Á, et al. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur Respir J 2011;37:617-23.
227. Aushev VN, Zborovskaya IB, Laktionov KK, et al. Comparisons of microRNA patterns in plasma before and after tumor removal reveal new biomarkers of lung squamous cell carcinoma. PLoS One 2013;8:e78649.
228. Wei F, Ma C, Zhou T, et al. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of miRNA-222-3p. Mol Cancer 2017;16:132.
229. Yuwen DL, Sheng BB, Liu J, Wenyu W, Shu YQ. MiR-146a-5p level in serum exosomes predicts therapeutic effect of cisplatin in non-small cell lung cancer. Eur Rev Med Pharmacol Sci 2017;21:2650-8.
230. Shi L, Chen J, Yang J, Pan T, Zhang S, Wang Z. MiR-21 protected human glioblastoma U87MG cells from chemotherapeutic drug temozolomide induced apoptosis by decreasing Bax/Bcl-2 ratio and caspase-3 activity. Brain Res 2010;1352:255-64.
231. Akers JC, Ramakrishnan V, Kim R, et al. MiR-21 in the extracellular vesicles (EVs) of cerebrospinal fluid (CSF): a platform for glioblastoma biomarker development. PLoS One 2013;8:e78115.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Tamura T, Yoshioka Y, Sakamoto S, Ichikawa T, Ochiya T. Extracellular vesicles as a promising biomarker resource in liquid biopsy for cancer. Extracell Vesicles Circ Nucleic Acids 2021;2:148-74. http://dx.doi.org/10.20517/evcna.2021.06
AMA Style
Tamura T, Yoshioka Y, Sakamoto S, Ichikawa T, Ochiya T. Extracellular vesicles as a promising biomarker resource in liquid biopsy for cancer. Extracellular Vesicles and Circulating Nucleic Acids. 2021; 2(2): 148-74. http://dx.doi.org/10.20517/evcna.2021.06
Chicago/Turabian Style
Tamura, Takaaki, Yusuke Yoshioka, Shinichi Sakamoto, Tomohiko Ichikawa, Takahiro Ochiya. 2021. "Extracellular vesicles as a promising biomarker resource in liquid biopsy for cancer" Extracellular Vesicles and Circulating Nucleic Acids. 2, no.2: 148-74. http://dx.doi.org/10.20517/evcna.2021.06
ACS Style
Tamura, T.; Yoshioka Y.; Sakamoto S.; Ichikawa T.; Ochiya T. Extracellular vesicles as a promising biomarker resource in liquid biopsy for cancer. Extracell. Vesicles. Circ. Nucleic. Acids. 2021, 2, 148-74. http://dx.doi.org/10.20517/evcna.2021.06
About This Article
Copyright
Data & Comments
Data
 Cite This Article 37 clicks
Cite This Article 37 clicks


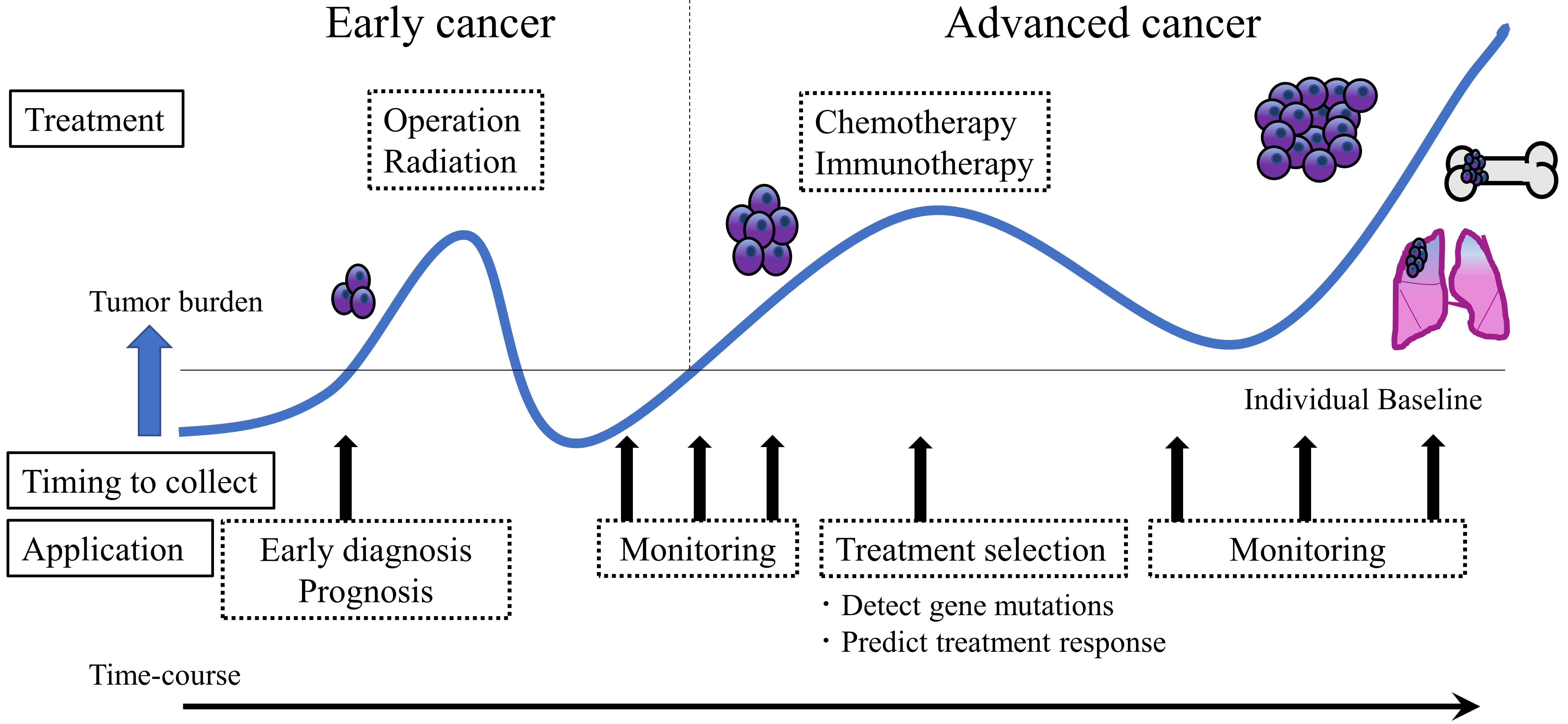
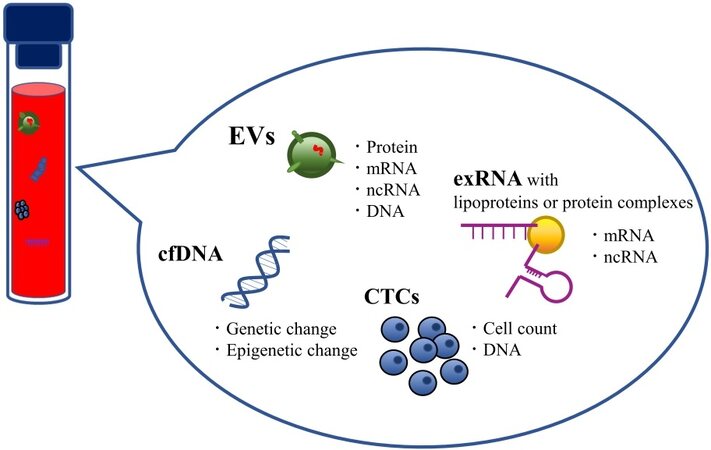











Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.