Extracellular vesicles: cross-organismal RNA trafficking in plants, microbes, and mammalian cells
Abstract
Extracellular vesicles (EVs) are membrane-enclosed nanometer-scale particles that transport biological materials such as RNAs, proteins, and metabolites. EVs have been discovered in nearly all kingdoms of life as a form of cellular communication across different cells and between interacting organisms. EV research has primarily focused on EV-mediated intra-organismal transport in mammals, which has led to the characterization of a plethora of EV contents from diverse cell types with distinct and impactful physiological effects. In contrast, research into EV-mediated transport in plants has focused on inter-organismal interactions between plants and interacting microbes. However, the overall molecular content and functions of plant and microbial EVs remain largely unknown. Recent studies into the plant-pathogen interface have demonstrated that plants produce and secrete EVs that transport small RNAs into pathogen cells to silence virulence-related genes. Plant-interacting microbes such as bacteria and fungi also secrete EVs which transport proteins, metabolites, and potentially RNAs into plant cells to enhance their virulence. This review will focus on recent advances in EV-mediated communications in plant-pathogen interactions compared to the current state of knowledge of mammalian EV capabilities and highlight the role of EVs in cross-kingdom RNA interference.
Keywords
INTRODUCTION
Extracellular vesicles (EVs) were first observed in plants and mammals in the mid-1900s, half a century before it would be known that these vesicles are a universally conserved and impactful form of inter-cellular and inter-organismal communication. The first mammalian observation of extracellular vesicles was made in 1946 when platelet-derived particles were detected in human plasma[1]. In the following decades, there were numerous scattered observations of mammalian EVs in vivo and in cell culture[2]. However, throughout the 1900s, these extracellular particles were often considered to be cellular debris, a term that historically suggested that these particles represented cellular waste[3]. This "debris" designation spurred a decades-long delay in the development of EV research, but since the turn of the century, this field has quickly expanded to show the diversity and influential functions of EVs on cellular development and interactions.
Research on plant- and microbial-derived EVs has undergone the same slow development as the study of mammalian EVs, although in recent years, there has been a surge of new studies characterizing EVs from plant and microbial cells, showing the same scope of diversity and biological significance as mammalian EVs. In cotton plants, intraluminal vesicles in multivesicular bodies were first observed in 1965[4]. This was followed by the observation in carrots that multivesicular bodies could fuse with the plasma membrane and release vesicles into the extracellular space, leading to the discovery of plant extracellular vesicles[5]. In the early 2,000s, studies of plant and microbial EVs using transmission electron microscopy revealed the accumulation of vesicles at plant-bacteria and plant-fungus interaction sites[6-8]. This inspired a new field of research into the role of plant and microbial EVs in the trafficking of materials not only between cells within an organism, but also between different organisms and kingdoms of life.
Numerous classifications and diverse roles of EVs have been characterized in mammalian systems. EVs can be isolated from plasma, serum, urine, and various other physiological fluids[9,10]. They are small lipid-encapsulated nanoparticles that carry cargoes, such as RNAs [e.g., small RNAs (sRNAs), messenger RNAs (mRNAs), transfer RNA (tRNA), and long non-coding RNAs], DNA fragments, lipids, and proteins[9,11-13]. EVs transport these biomolecules between cells and play an important role in cell-to-cell communication[9,14]. In addition, EVs have emerged as important disease biomarkers and have potential applications in therapeutics and diagnostics[15-17]. Among all cellular origins, including mammalian, plant, and microbial cells, EVs can be broadly classified into either exosomes or microvesicles based on their biogenesis[3,10,18]. Exosomes are small EVs (30-150 nm in diameter) originating from the endosomal organelle multivesicular bodies (MVBs). Microvesicles, also denoted as ectosomes, are heterogeneous and larger EVs (100-1,000 nm in diameter) generated by direct budding from the plasma membrane[19,20]. In contrast to exosomes and microvesicles, mammalian cells also produce several types of vesicle bodies associated with different forms of cell death[3]. These include necroptotic vesicles released from necroptotic cells[21,22], pyroptotic inflammasomes released during pyroptosis of cells[23], and apoptotic bodies (1-5 µm in diameter) produced during apoptotic cell disassembly[24-26]. Research on EVs from plants and microbes has progressed less than research in mammalian systems due to the limitations in EV isolation and detection methods[27]. Mammalian EVs are frequently isolated from cell cultures or body fluids such as blood, breast milk, urine, lung fluid, and semen[2,28]. Since these mammalian samples are fluids, extraction is relatively straightforward. In contrast, extraction of EVs from plants is more challenging due to the small amount of extracellular fluid and the extensive procedures required for EV isolation in plants.
Recently, plant EVs have been successfully isolated from the apoplastic fluids of leaves, roots, and seeds[29-34]. Research has attempted to characterize different classes of plant EVs, protein markers, and lipid composition, mirroring the work done to characterize mammalian EVs. Emerging evidence indicates that plant EVs play an essential role in the cross-kingdom transport of plant sRNA into fungal cells to silence virulence-related genes[29,35,36]. This phenomenon, termed "cross-kingdom RNAi", has been observed in interactions between numerous plant and animal hosts with their associated microbes and parasites[35,37-41]. In some host-pathogen/parasite interactions, RNA translocation has been linked to EV trafficking[29,42-46]. The wide range of organisms capable of RNA exchange highlights the importance of research into the mechanisms of RNA translocation and the potential role of EVs in these interactions. Recent research has also shown that for plant-infecting microbes such as bacteria, fungi, and oomycetes, EVs could play a prominent role in pathogenesis by delivering proteins, RNAs, and metabolites into their plant host cells[47-52] [Figure 1]. In this review, we highlight and discuss the current state of research on plant-derived EVs and cross-kingdom RNAi, with an emphasis on the role of EVs in cross-kingdom RNA trafficking. We also discuss plant-interacting pathogen-derived EVs and their biological functions. Finally, we include the potential applications of plant EVs for agricultural crop protection and advances in human medicine.
Figure 1. Cross-kingdom RNAi in plant–microbial interactions. Some fungal pathogens, such as B. cinerea and V. dahlia, deliver sRNAs into the plant cells to silence host genes that are involved in plant immunity[88,91]. Cross-kingdom RNAi is bidirectional, and plants secrete TET8/9-positive EVs to transport host functional sRNAs into pathogens to silence fungal genes involved in virulence[29]. TET8/9-positive EVs contain a variety of RBPs, including AGO1, RHs, and ANNs, which contribute to the selection or stabilization of sRNAs in EVs[56]. Cross-kingdom RNAi also exists in bacteria-plant interaction. Rhizobia tRNA-derived short fragments act as functional sRNAs moving into plant cells to silence target genes related to nodulation[113]. sRNAs from fungal pathogen and bacterium rhizobia were all found to be loaded into plant host AGO1 to silence host target genes. Fungi and bacteria are predicted to secrete and transport sRNAs into host cells by EVs. The question mark indicates a prediction that has not yet been validated experimentally. EVs: extracellular vesicles; MVB: multivesicular body; ILV: intraluminal vesicle; OM: Outer membrane; PM: plasma membrane; sRNA: small RNA; TE: Transposable element; tRFs: tRNA-derived fragments.
PLANT EXTRACELLULAR VESICLES
EVs provide protection for their biological cargo from the abundant nucleases and proteases within the extracellular environment[53,54]. EV-mediated trafficking is one of the major pathways for RNA transport. Mammalian EVs can be isolated from the extracellular environment of biological fluids or culture supernatants[9,10]. Similarly, plant EVs from various sources can be isolated from extracellular apoplastic washing fluid and cell culture medium[29,30] [Figure 2]. Plant EVs have been observed in numerous plant species such as cotton[4], carrot[5], and rice[55] and have been isolated from Arabidopsis thaliana[29-31,56], sunflower[32,57], olive[58,59], and Nicotiana benthamiana[56,60] [Figure 2].
Figure 2. Heterogeneous populations of EVs isolated from plants. There are at least four known EV populations that have been isolated from plants: TET-positive EVs, PEN1-positive EVs, autophagy-related EVs, and pollenosomes. They have different sizes, densities, cargoes, and intracellular origins. Pathogen infection induces secretion of both TET-positive EVs and PEN1-positive EVs[56]. In the process of EV isolation, final centrifugation of 40,000 × g (P40) pellets larger and heavier vesicles such as PEN1-positive EVs and large EVs, non-vesicular free RNA, and RNA-protein complexes[30,79]. Small EVs, such as TET-positive EVs, are mainly present in the supernatant after 40,000 × g centrifugation and require a higher speed of ultracentrifugation at 100,000 × g for collection (P100)[30,56]. Autophagy-related EVs marked with ATG8a were collected using 100,000 × g from plants during the autophagy process within cells[64]. Pollenosomes secreted during pollen germination and pollen tube growth were collected at 100,000 × g from in-vitro pollen gemination media[59]. EXPO-derived EVs originate from the plant-specific organelle EXPO, are marked by the protein Exo70E2, and have not yet been isolated from plants[71]. This figure was created with https://www.biorender.com/.
Heterogeneous EV groups have been isolated in plants, which may perform different functions[61,62]. In Arabidopsis, five EV subtypes have been reported that are either isolated and/or characterized by different protein markers, sizes and/or biogenesis origins, as described in Figure 2. These subtypes include tetraspanin (TET)-positive EVs[29,56], penetration 1 (PEN1)-positive EVs[63], exocyst-positive organelle (EXPO)-derived EVs[45], autophagy-related EVs[64], and pollensomes[59]. TET-positive EVs are the class of plant EVs derived from MVBs and most similar to mammalian exosomes. In Arabidopsis, the secretion of TET8-positive EVs increases after the infection of fungal pathogen Botrytis cinerea[29,56]. These plant exosomes are enriched in the supernatant after centrifugation of leaf apoplast fluid at 40,000 × g (P40 fraction), and can be collected by centrifugation at 100,000 × g (P100-P40), the same speed used to collect mammalian exosomes. Plant exosomes are mainly responsible for transporting sRNAs, including microRNAs (miRNAs) and small interfering RNAs (siRNAs), from plants to fungal pathogens to induce cross-kingdom RNAi[29,56]. TET8 is a homolog of the mammalian EV-enriched tetraspanin proteins CD9, CD63, and CD81. The CD9-, CD63-, and CD81-labeled mammalian EVs are generated from MVBs[65]. In mammalian cells, Rab GTPases are essential for intracellular vesicle movement and strongly influence MVB morphology[9]. TET8-labeled plant EVs colocalize with the Arabidopsis MVB-marker Rab5-like GTPase ARA6 and accumulate at fungal infection sites, suggesting that TET8-positive EVs are likely originated from MVB trafficking[29,56]. Similarities between plant and mammalian exosomes suggest a conserved mechanism in exosome biogenesis between mammalian and plant cells. In mammals, ceramide plays an important role in exosome biogenesis and release[66]. Interestingly, sphingolipids in Arabidopsis EVs are nearly pure glycosyl inositol phosphoryl ceramides (GIPCs)[67]. GIPCs are only produced by plant and fungal species and are not present in mammalian EVs[68]. The amount of cellular GIPCs is lower in the tet8 knockout mutant than in the wild type, and the tet8 mutant secretes fewer EVs[67]. Exogenous application of GIPCs can rescue the EV secretion defects in the tet8 mutant[67], illustrating the important role of TET and GIPCs in plant EV formation.
Less is known about the additional types of plant EVs, such as PEN1-positive EVs, EXPO-derived EVs, and autophagy-related EVs. PEN1 is a plant-specific plasma membrane-associated syntaxin protein[69]. The biogenesis pathway of PEN1-positive EVs is currently unclear. PEN1-positive EVs are mostly enriched in the P40 fraction and can be collected by centrifugation of apoplastic fluid at 40,000 × g. They mainly carry tiny RNAs with 10-17 nt in length[63]. The potential destination and biological function of these tiny RNAs remain to be investigated. EXPO-derived EVs originate from the plant-specific novel organelle EXPO, which is marked in plants by the exocyst protein Exo70E2[70,71]. EXPO is independent of endosomes and autophagosomes[71]. EXPO-derived EVs are a class of large EVs with diameters ranging from 200-500 nm, larger than exosomes[70,71]. To date, EXPO-derived EVs have not been isolated and their specific cargoes and biological functions also remain unclear. Autophagy-related vesicles have recently been shown to be released as EVs at the late stage of pathogen infection[64]. Within cells, these autophagosomes colocalize with monolignol metabolites and are implicated in the transport of monolignols across the plant plasma membrane into the apoplast to facilitate lignin deposition as part of the plant defense response to pathogens[64]. Similar to mammalian systems, plant EVs have been isolated from various tissue types throughout the organism[45]. Small plant EVs were isolated from the media used for in vitro pollen germination and pollen tube growth[58,59]. These small EVs are termed pollensomes, and their size ranges from 28 to 60 nm in diameter[59]. In addition to EVs being a conserved structure among different mammalian species, EVs are currently being discovered in many plant species. Rice EVs have recently been observed in the periarbuscular space after inoculation with the symbiotic arbuscular mycorrhizal fungi Rhizophagus irregularis or Gigaspora rosea using transmission electron microscopy[55]. Whether these EVs carry RNA requires further study.
It is worth noting that not all plant-derived vesicles are EVs. EVs are specifically and naturally secreted vesicles found in the extracellular space of plants and have distinct biomarkers and cellular functions[30,45]. Previously, vesicles collected from disrupted or homogenized tissues of grape, broccoli, ginger, and grapefruit were termed nanovesicles[72,73]. However, these nanovesicles are not true EVs, because they do not naturally occur in extracellular spaces. Nevertheless, plant nanovesicles have biomedical applications because they can protect and deliver biological compounds, such as drugs, RNAs and proteins, into target cells[74]. Plant nanovesicles play a role in the prevention of inflammation and intestinal permeability in humans and mice and influence the intestinal microbiome[75]. Consistent with their separate classifications, plant nanovesicles and Arabidopsis-derived EVs have different size distributions: EVs range from 60-200 nm, and nanovesicles range from 100-300 nm[76]. Cancer cells take up both types of vesicles with high efficiency, although EVs have an almost three-fold higher uptake rate than plant nanovesicles[76]. This result highlights the functional diversity of plant-derived vesicles and supports the idea that plant EVs have the natural property of transporting cargo between cells. These results suggest that plant EVs have potential applications in the delivery of therapeutics[77,78].
COMMONLY USED METHODS TO EXTRACT PLANT EXTRACELLULAR VESICLES
For mammalian cells, EVs can be isolated from extracellular fluids using differential centrifugation to obtain different subgroups of EVs with different density ranges[9]. For instance, low-speed centrifugation (around 2,000 × g) can effectively recover large vesicles or cell fragments like apoptotic bodies. Intermediate-size EVs, such as microvesicles, can be collected by centrifuging at around 10,000-20,000 × g. Small EVs, predominantly exosomes, require high-speed ultracentrifugation over 100,000 × g (P100) for successful recovery. In plants, two ultracentrifugation speeds have been used to pellet EVs: 40,000 × g, which collects the P40 fraction, and 100,000 × g, which collects the P100 fraction. For plant EVs pelleted at 100,000 × g, 92% have diameters in the range of 30-150 nm[30]. Mammalian EVs pelleted at the same ultracentrifugation speed have similar diameters below 150 nm and are denoted as exosomes[9,10,61]. In plants, the P100 fraction is enriched in the exosome-like TET8-positive EVs, which contain sRNAs and RNA-binding proteins[79]. Most TET8-positive EVs remain in the P40 supernatant, and centrifugation of the supernatant of P40 at 100,000 × g (the P100-P40 fraction) collects mostly TET8-positive EVs[79] [Figure 2]. The P40 fraction is enriched in PEN1-positive EVs, which contain tiny RNAs and proteins[79]. As well in the P40 fraction, non-vesicular sRNAs and circular RNAs associated with RNA binding proteins co-pellet with EVs[79]. While circular RNAs are more resistant to degradation because of lacking free ends, it is currently unknown how other liner non-vesicular RNAs, including sRNAs, could be protected from degradation within the plant apoplast that contains numerous nucleases[53,54], and whether RNA binding protein alone can completely protect the non-vesicular RNAs from degradation. These studies using different ultracentrifugation speeds for plant EV isolation are complementary to the discovery and characterization of distinct classes of EVs with different sizes, densities, and cargoes.
In tandem with different centrifugation speeds, differences in the extraction methods of apoplastic washing fluid can greatly affect the collection of different EVs or non-vesicular contents. Studies characterizing the P40 fraction notably isolated EVs and non-vesicular RNAs/RNA-protein complexes from apoplastic washing fluid extracted from the whole plant, rather than detached leaves[31,79]. Apoplastic washing fluid collected by the whole plant method contains more contamination from cell debris and cytoplasmic content compared to the extraction of apoplastic washing fluid from the detached leaves method[30]. This is evident through Western blotting analysis, which detected a high amount of Rubisco protein in EV fractions isolated with the whole plant method[30]. In comparison, the detached leaves method resulted in minimal Rubisco protein contamination in EV preparations[30]. It is important to consider not only extraction methods, but also the disease or stress conditions of the plant that affect EV cargoes as well as non-vesicular contents that co-pellet with EVs. Different methods may only capture different subsets of EVs, and it is important to avoid generalizations about the function of all plant EV subtypes based on individual methods.
Differential centrifugation followed by gradient centrifugation can help increase the quality of isolated EVs and reduce contaminations[29,30]. Gradient centrifugation can be done using a sucrose gradient[80] or an iodixanol gradient[31]. Different plant EV subtypes are found in different density fractions, which has been shown through the detection of EV protein markers[30,31]. PEN1-positive EVs are enriched in the iodixanol gradient fraction from 1.029 to 1.056 g/mL[31]. TET8-positive EVs are enriched in the iodixanol fraction of 1.08 g/mL[30], which is similar in density to mammalian exosomes (1.08-1.12 g/mL)[81,82]. Importantly, TET8-positive EVs are in the same fractions of plant miRNAs and siRNAs[56], suggesting that TET8-positive EVs are the major class of EVs for sRNA transport. Along with pelleting at different final ultracentrifugation speeds, enrichment in different gradient fractions further suggests that TET8‐positive EVs and PEN1‐positive EVs represent two distinct subpopulations with different densities. Furthermore, in transgenic plants co-expressing fluorescently labeled TET8 and PEN1, TET8-GFP-labeled EVs and PEN1-mCherry-labeled EVs do not colocalize, supporting the distinction between these two classes of EVs[30,56]. Differential centrifugation, sucrose gradients, and buoyant density gradients also reduce the occurrence of apoptotic cell fragments in the final pellet[83].
Methods for isolation of plant EVs, such as differential centrifugation and gradient density centrifugation, can fractionate different classes of EVs based on size and density, but these methods cannot sufficiently purify one specific EV subtype in isolation. The method that is able to purify one specific EV class is immunoaffinity purification using an antibody that recognizes a specific protein marker of the particular EV class. A TET8 native antibody that recognizes an extracellular domain 2 (EC2) of TET8 has been successfully generated to isolate plant TET8-positive EVs[30,56]. Plant miRNAs and siRNAs were detected in immunoaffinity purified TET8-positive EVs[56], supporting that plant sRNAs are mainly transported by exosomes. PEN1-positive EVs and EXPO-derived EVs have not been successfully separated from heterogeneous EV classes. The preparation of antibodies that recognize the extracellular domains of PEN1 and EXO70E2 to purify PEN1-positive EVs and EXPO-derived EVs will help determine the RNA cargoes and other molecules in these subclasses of EVs. It is highly desirable that immunoaffinity capture be used to properly assess specific cargo of different vesicle classes. Recent technical advances in flow field-flow fractionation chromatography may make it possible to sort different classes of EVs by different sizes or fluorescence labeling[84].
CROSS-KINGDOM RNAI
sRNAs are short non-coding regulatory RNAs that silence genes with complementary sequences[37,85]. In eukaryotes, sRNAs are generated by the ribonuclease III-like enzyme Dicer or Dicer-like (DCL) proteins. sRNAs are then incorporated into Argonaute (AGO) proteins to form an RNA-induced silencing complex, which performs gene silencing in a sequence-specific manner by mRNA cleavage and degradation, translational inhibition, or transcriptional gene silencing. During microbial infection, host RNAi machinery is critical for reprogramming gene expression to induce plant immunity[37,86,87]. Recent studies demonstrate that, in addition to their endogenous functions, sRNAs travel between hosts and their interacting organisms and induce ‘cross-kingdom or cross-organismal RNAi’ in trans[35,38,88-90] [Figure 1].
Cross-kingdom RNAi was first observed in the interaction between Arabidopsis and the pathogen Botrytis cinerea[91]. During infection on both Arabidopsis and tomato host plants, B. cinerea delivers sRNAs into plant cells that silence plant immunity genes by binding to the host AGO1 protein[91,92]. Pathogen sRNAs associate with host AGO proteins and target genes involved in plant immune responses, including plant kinase genes, which are often involved in signal transduction pathways that mediate plant defense [93]. B. cinerea sRNAs specifically target mitogen-activated protein kinase (MAPK) pathway genes and cell wall-associated kinases[91]. Like B. cinerea, sRNAs from the fungal wilt pathogen Verticillium dahliae also bind to Arabidopsis AGO1 for host gene silencing[88,94]. In the fungal rust pathogen of wheat, Puccinia striiformis, a micro-like RNA (milRNA) serves as an important pathogenicity factor that silences pathogenesis-related 2 (pr2) gene, encoding a β-1,3-glucanase, which is a key wheat immunity gene[95]. As well, other P. striiformis sRNAs are predicted to target wheat kinase genes[96]. On tomato hosts, the fungal pathogen Fusarium oxysporum generates mil-R1, which is loaded into tomato AGO4 to silence the tomato CBL‐interacting protein kinase gene[97]. In addition to fungal pathogens, the oomycete pathogen Hyaloperonospora arabidopsidis sends sRNAs into Arabidopsis cells that associate with Arabidopsis AGO1 to silence defense-related genes: with no lysine kinase (AtWNK)2 and apoplastic, enhanced disease susceptibility-dependent (AtAED)3[98]. sRNAs from the biotrophic powdery mildew pathogen, Blumeria hordei, have also been predicted to target possible barley host immunity-related genes[99].
Fungal sRNA effectors play an important role in fungal pathogenicity and virulence. The majority of fungal sRNA effectors are derived from long terminal repeat (LTR) retrotransposons and are generated by fungal DCLs (DCL1 and DCL2)[91]. Mutation or silencing of fungal DCL genes can attenuate the virulence and growth of fungal pathogens. Mutation of both DCL genes reduced the virulence and growth of the grey mold pathogen B. cinerea[88,91,100], the soilborne pathogen Fusarium graminearum[101], the post-harvest decay pathogen Penicillium italicum[102], the apple canker pathogen Valsa mali[103], and the grapevine downy mildew pathogen Plasmopara viticola[104]. Furthermore, even silencing of DCL genes, reducing expression of both DCL genes without knocking out expression, resulted in reduced pathogenicity of B. cinerea[88,100,105], F. graminerum[106], and P. viticola[104]. Mutation of DCL genes has also shown reduced virulence and growth rate of the anthracnose pathogen Colletotrichum gloeosporioides[107]. TEs are major components of eukaryotic genomes and give rise to many siRNAs[89,108]. In phytopathogens, the LTR retrotransposons are associated with virulence in host-adapted subpopulations of B. cinerea[109]. This phenomenon may be due to sRNA effectors being generated from these LTR regions. Thus, cross-kingdom transported sRNAs originating from these TEs could explain the rapid variation of sRNA species produced during the co-evolutionary arms race with the host.
Beyond fungal pathogens and plants, cross-kingdom RNAi is a conserved mechanism across many diverse species. The parasitic plant dodder Cuscuta campestris delivers 22-nucleotide miRNAs into Arabidopsis, which can act as virulence factors during parasitism[110]. Cross-kingdom RNA trafficking also occurs in host-symbiontic interactions. For example, to stabilize a symbiotic interaction, the ectomycorrhizal fungus Pisolithus microcarpus transports miRNAs into Eucalyptus grandis cells to silence host NB-ARC domain-containing genes. This attenuates the superfluous immune response of the plant to accommodate the symbiont[111]. As well, in the symbiosis between the arbuscular mycorrhizal fungi Rhizophagus irregularis and its host plant Medicago truncatula, fungal sRNAs are predicted to target host mRNAs of genes whose expression is modulated in roots after mycorrhizal colonization[112]. Furthermore, sRNA trafficking between hosts and microbes is not limited to eukaryotes with RNAi machinery. The plant symbiotic bacterium Rhizobium transports 21-nucleotide tRNA-derived sRNA fragments (tRFs) into its soybean host, which are loaded into soybean AGO1 and cleave nodulation-related mRNAs, contributing to the symbiotic interaction[113]. It remains to be determined whether cross-kingdom transport of tRFs into plant hosts occurs during pathogenic bacterial infections.
Cross-kingdom RNAi is bidirectional, in which sRNAs are sent from the microbe into the host and also from host cells into interacting microbes. As part of the plant defense response, some plant sRNAs, generated by DCL proteins[114], are transported into interacting microbes/pests and silence target genes necessary for pathogen/pest infection, thereby reducing plant disease[38]. In Arabidopsis, trans-acting small interfering RNAs (tasiRNAs) are trafficked into interacting B. cinerea cells and silence fungal vesicle trafficking-related genes essential for virulence, such as vacuolar protein sorting 51 (VPS51), dynactin (DCTN1) and suppressor of actin(SAC1)[29]. The transport of host plant endogenous sRNAs into pathogens has been observed in other pathosystems. Tomato plants transport multiple miRNAs into B. cinerea cells that attenuate virulence by suppressing spore germination and silencing virulence-related genes[115,116]. Cotton plants produce miR159 and miR166 that respectively target genes in V. dahlia that are essential for fungal virulence[117]. As well, wheat plants produce a miRNA that silences the expression of a hydrolase enzyme in the fungus Fusarium graminearum which directly decreases its virulence[118].
During the arms race between hosts and pathogens, some pathogens have also developed countermeasures to suppress cross-kingdom RNAi. A recent study showed that the fungal pathogen V. dahliae has an adaptive mechanism to evade host-derived cross-kingdom RNAi. V. dahliae secretes protein secretory silencing repressor 1 (VdSSR1) into plant cells, which interferes with host AGO1-miRNA export from the nucleus and inhibits antifungal cross-kingdom RNAi[119]. In addition,to promote infection, the oomycete Phytophthora capsici produces an effector called Phytophthora suppressor of RNA silencing 2 (PSR2), which inhibits the biogenesis of Arabidopsis secondary siRNAs that target Phytophthora genes via cross-kingdom RNAi[43].
Beyond plant-microbial/parasite interactions, cross-kingdom RNAi or cross-organismal RNAi has also been observed between mammalian and insect hosts and their pathogens/parasites. In insects, the fungal pathogen Beauveria bassiana transports a milRNA into mosquito cells and uses the same mechanism as the plant fungal pathogen by hijacking mosquito host AGO1 to silence the expression of a mosquito Toll receptor ligand, thereby attenuating mosquito immunity[120]. Conversely, in this interaction, B. bassiana infection induces the expression of two mosquito miRNAs, let-7 and miR-100. These miRNAs move into the fungal cells and specifically silence the sec2p and C6TF fungal genes, both of which are essential for fungal growth and pathogenicity[121]. In mammals, the gastrointestinal nematode Heligmosomoides polygyrus transports miRNAs into mouse intestinal epithelial cells to suppress innate immune responses and eosinophilia[42]. As well, human monocyte cells export miRNAs into the fungal pathogen Candida albicans to silence the cyclin-dependent kinase inhibitor Sol1[122]. Human and mouse gut epithelial cells also secrete miRNAs that can regulate gene expression and growth of gut bacteria, such as Fusobacterium nucleatum and Escherichia coli[44]. Although bacteria do not possess RNAi machinery, bacterial endoribonucleases may mediate this cross-kingdom gene regulation. Alternatively, host sRNAs could be transported together with host AGO proteins to silence bacterial genes in trans. There have also been reports about the possible functions of dietary sRNAs and their potential role in modulating endogenous gene expression in mammals. Although different studies using different systems with varying experimental conditions have produced contradictory results, this nascent area of research into the therapeutic effects of dietary sRNAs is worth pursuing[123]. These observations suggest that cross-kingdom RNAi occurs throughout many branches of life between various parasites/microbes and their hosts.
EXTRACELLULAR VESICLES IN CROSS-KINGDOM SMALL RNA TRAFFICKING
Mammalian exosomes from different cell types have been found to carry different RNAs, such as miRNAs, mRNAs, and long non-coding RNAs, throughout the body for various functions in disease, cancer, and immune responses[3,17]. For example, human breast cancer cells produce EVs containing miRNA precursors and DICER and AGO2 proteins that function together to perform gene silencing in naive non-cancerous cells[124]. Human T-cells also produce exosomes with specific exosome-enriched miRNAs that are transported into infected antigen-presenting cells to modulate their gene expression as part of the immune response[125]. Renal cancer cells secrete EVs loaded with a long non-coding RNA that acts in tandem with miRNAs in cells to modulate gene expression and overcome the effects of anticancer drugs[126]. Cancer-derived EVs carrying intact mRNAs also facilitate the spread of ovarian cancer into the peritoneal cavity[127]. There is a much greater currently characterized diversity of types and functions of RNAs in mammalian EVs compared to plant or microbial EVs.
EVs can be isolated from uninfected healthy plants[29,31] but are more enriched in plants after pathogen infection or treatment with the plant defense hormone salicylic acid[29,31,56], suggesting that EVs play an important role in plant immunity. Comparative analysis on sRNA profiles of purified fungal cells and EVs isolated from the infected tissue show that more than 70% of transferred plant sRNAs found in fungal cells could be detected in plant EVs pelleted at 100,000 × g[29]. This suggests that EVs are one of the major pathways for cross-kingdom sRNA trafficking. Using density gradient analysis and direct immunoaffinity capture, EV-enriched sRNAs are present in the fractions of TET8-positive EVs[56]. Furthermore, double mutants of Arabidopsis tet8 and its close homolog tet9 (tet8/tet9) deliver much fewer sRNAs into fungal cells and show increased susceptibility to B. cinerea infection[29]. These biochemical and genetic results further support that TET8-positive EVs are the major class of EVs that transport sRNAs to fungal cells
EV-mediated sRNA trafficking is also observed between mammalian hosts and their pathogens and parasites, demonstrating the important role of EVs in cross-organismal sRNA trafficking. In the aforementioned interaction between the nematode H. polygyrus and mice, H. polygyrus uses EVs to transport miRNAs into the mouse cells[42]. Similarly, human monocyte cells that deliver miRNAs into fungal pathogen C. albicans also rely on EVs for this cross-kingdom RNAi[122]. Within intra-organismal cellular interactions in mammalian systems, EVs have been shown to traffic functional mRNA transcripts between cells for protein translation in destination cells[128,129]. Human and mouse white blood cells produce EVs containing hundreds of functional mRNA species[128]. mRNA trafficking was a novel discovery in mammalian cell-cell communication and revealed a new function of EV trafficking[127]. Despite mRNA intra-organismal trafficking via EVs, there have been no reports to date of functional mRNA cross-kingdom trafficking in mammals, plants, or microbes.
SELECTIVE LOADING OF SMALL RNAs INTO EVs
Selective sorting of sRNAs into EVs has been previously reported and continues to be characterized in many mammalian cell types. In human lymphocytes, hnRNPA2B1 protein was found to bind to sRNAs and load sRNAs into exosomes depending on specific sequence motifs[130]. A cleaved form of Rab-interacting lysosomal protein (RILP) also contributes to exosomal miRNA loading depending on a conserved RNA motif (AAUGC) in cultured HeLa cells[131]. Another study in mouse neurons showed that miRNAs that share a conserved four-nucleotide motif (GUAC) were selectively loaded into EVs[132].
Although selective RNA loading into EVs based on conserved motifs or RNA-binding proteins is established in mammalian vesicle biology, research on selective loading is just beginning to be explored in the study of plant and microbial EVs. In Arabidopsis, EV-sRNA profiling analysis revealed that the enrichment of sRNA species in EVs does not correlate with their abundance inside the plant cell[56], which suggests that these EV-enriched sRNAs are selectively loaded into EVs for transport. Proteomics analysis on purified EVs isolated from B. cinerea infected Arabidopsis leaves revealed several RNA-binding proteins, including Arabidopsis AGO1, DEAD-box RNA helicases (RH) 11, RH37, and RH52, which contribute to EV selective sRNA loading[56]. These RNA-binding proteins are localized in the membrane fractions inside the cell and selectively bind with a subset of sRNAs that are 20-22 nt long, with the first 5’ nucleotide being uracil. These RNA-binding proteins subsequently enter MVBs with associated sRNAs and are released within exosomes into the apoplast. Another set of EV-localized RNA-binding proteins Annexin (ANN) 1 and ANN 2 bind sRNAs non-specifically. Furthermore, ago1, rh11/37, and ann1/2 mutants have reduced levels of EV-enriched sRNAs in EVs[56]. These results demonstrate that AGO1 and RH11/37/52 are important for selective sRNA loading into EVs, and that although ANN1/2 are not involved in the selective loading process, they are important for sRNA stabilization in EVs [Figure 1][56]. Similar EV-mediated cross-species RNAi and RNA loading mechanisms exist in mammalian-parasite interactions. EVs of nematode parasite H. polygyrus, which mediate cross-species RNAi within its mammalian host[42], contain an AGO protein (exWAGO) that specifically binds to EV-enriched RNAs[133]. Taken together, these results indicate that AGO proteins are one of the components that are mainly responsible for selective RNA sorting into EVs in plants and nematodes.
PATHOGEN-DERIVED EXTRACELLULAR VESICLES AND THEIR BIOLOGICAL FUNCTIONS
EV secretion is a conserved process in all eukaryotic and prokaryotic cells[10,45]. Research has begun to demonstrate the important role of microbial EVs in mediating the ability of bacteria and fungi to interact with their hosts. Microbes, such as Gram-positive and Gram-negative bacteria, archaea, and fungi, have all been shown to release EVs[134,135]. Gram-negative bacteria release 50 to 300 nm EVs by pinching off the bacterial outer membrane, generating outer membrane vesicles (OMVs)[136-138]. It has been debated how vesicles can be released or taken up by organisms with thicker cell walls compared to the ease of vesicle movement within mammalian cells lacking cell walls. A recent review highlighted the plasticity of the fungal cell wall, allowing for the passage of vesicles as large as 200 nm in and out of fungal cells, despite the original measurement of fungal cell wall pores being much smaller[139,140]. Recent proteomics data showed that fungal EVs are enriched with cell wall-modifying enzymes[50,141], which could allow for movement of fungal EVs through the plant cell wall. This emphasizes the dynamic and flexible nature of the cell wall and provides information on how vesicles could move in and out of the cells of thicker-walled organisms. As well, EV membranes are malleable and have sufficient structural plasticity to facilitate passing through the cell walls.
Microbial EVs contain specific cargoes, such as DNA, RNA, metabolites, and proteins, that could be involved in pathogenesis and modulation of host immunity[135,137,141,142]. Most microbial EVs are isolated from cellular cultures by differential centrifugation at a predetermined time point to avoid capturing dying cells[134,141,143]. Bacteria in the human gut have been shown to release bacterial EVs that can enter into the bloodstream and lymphatic system and travel to distant organs throughout the body[144]. Bacterial EVs can have beneficial or detrimental effects on human cells, such as inducing dendritic cell polarization or stimulating cancer proliferation[144]. The human fungal pathogen Cryptococcus neoformans also produces EVs that contain components of its fungal capsule, a cellular structure essential for virulence[145]. Proteomics analysis of EVs from five species within the genus Candida identified 36 common proteins enriched for orthologs of biofilm mediators of the mammalian pathogen C. albicans[146]. This study also discovered that vesicles from one Candida species can confer function to other Candida species, suggesting an important role of EVs in the development of microbial biofilm communities[146]. Scanning electron microscopy and cryo-transmission electron microscopy allowed the observation of EVs with an average size of 100 nm on the surface of the Candida biofilm, corroborating that EVs can easily move across the fungal cell wall[146].
Over the past decade, studies of microbial EVs have mainly focused on mammalian-infecting pathogens, with limited reports on EVs derived from plant pathogens. To date, most of the plant-interacting bacteria from which EVs were isolated are Gram-negative bacteria, including Xanthomonadaceae family bacteria[147-150], Xylella fastidiosa[151,152], and Pseudomonas syringae[153-155]. Proteomic analysis of Gram-negative bacterial EVs reveals virulence factors and signaling molecule contents that may play a role in plant infection[143]. Direct evidence that phytopathogen-derived EVs contribute to virulence was provided by the study of the xylem-colonizing plant pathogenic bacterium, X. fastidiosa[152]. X. fastidiosa-derived EVs block X. fastidiosa cells from interacting with the xylem wall, which increases its systemic spread within the plant host and promotes virulence[152]. It has also been observed that animal pathogenic bacteria produce EVs that fuse with the host plasma membrane[156]. The opportunistic human pathogen Pseudomonas aeruginosa produces EVs that fuse with the host cell plasma membrane at lipid raft microdomains for long-distance delivery of bacterial virulence factors[157]. Similarly, the phytopathogen Xanthomonas campestris generates EVs that fuse directly with the Arabidopsis plasma membrane. This process depends on the clustering of the plant lipid raft proteins remorin 1.2 and remorin 1.3[150]. The observation that phytobacterial EVs fuse with the host plasma membrane suggests that EVs facilitate cross-kingdom delivery of pathogen cargo to plant cells. However, whether phytobacterial EVs also contain RNA or DNA molecules is still unknown. As indicated above, symbiotic Rhizobium bacteria transport tRFs into host root cells to silence host nodulation genes, suggesting that bacterial tRNA may act as an sRNA source to mediate cross-kingdom RNAi[113]. It is interesting to pose whether Rhizobium RNA trafficking is mediated by EVs [Figure 1].
More than twenty species of yeast and filamentous fungi have been observed to secrete EVs[141]. Fungal EVs are thought to be derived from MVBs or to bud directly from the plasma membrane[135,142]. In the phytopathogenic fungus Ustilago maydis, five effectors and two membrane proteins form a stable protein complex anchored in EV-like structures that may be derived from MVBs[158]. In plant pathosystems, EVs have been isolated from several filamentous fungi, including Fusarium oxysporum[51], Fusarium graminearum[49], wheat pathogen Zymoseptoria tritici[50], corn smut fungus Ustilago maydis[159], anthracnose pathogen Colletotrichum higginsianum[160], post-harvest rot pathogen Penicillium digitatum[47], and powdery mildew fungus Blumeria hordei[99]. These EVs were purified from culture supernatants or fungi grown on plant tissue and contained proteins, nucleic acids, lipids, metabolites, and polysaccharides. Proteomic analysis of F. oxysporum EVs detected functional proteins involved in secondary toxin metabolism, cell wall degradation, and protein degradation[51]. Furthermore, injection of F. oxysporum EVs into the leaves of cotton or N. benthamiana plants induced a phytotoxic response in plants, suggesting that fungal EVs likely play an important role in infection[51]. In addition, critical virulence-related proteins and effectors have been identified in fungal EVs. For instance, proteomic analysis of F. graminearum EVs identified protein effectors, some of which do not have predicted secretion signal peptides[49]. This suggests that EVs are an unconventional secretion pathway for fungal effectors. EVs isolated from the fungus Z. tritici also contain a number of putative virulence-associated proteins that may play a role in the infection of wheat[50]. Current phytopathogen effector analysis focuses exclusively on predicted secreted proteins with signal peptides. Proteomics analysis of pathogen EVs may help identify a novel class of previously unidentified pathogen effectors.
Studies of RNA in EVs from plant pathogenic fungi are limited. EVs from U. maydis contain mRNAs encoding metabolic enzymes, known effectors, and virulence proteins[159]. EVs isolated from the infection site of Blumeria hordei on barley plants were enriched in B. hordei-derived milRNAs, which have a potential role in host gene silencing[99]. As mentioned above, the fungal pathogens B. cinerea, V. dahliae, and the oomycete pathogen H. arabidopsidis have been shown to deliver sRNAs into plant host cells[88,91,98]. The mechanism of the delivery of these sRNAs is likely via fungal EVs [Figure 1], and many experiments have been initiated to test this hypothesis. Aside from RNA and proteins, one study found that P. digitatum uses EVs to transport phytotoxic metabolites into citrus fruit cells during infection[47]. This is one of the first characterized reports of cross-kingdom metabolite trafficking in a host-pathogen interaction, demonstrating the functional diversity of EVs and their potential to traffic a variety of biological cargoes that have yet to be discovered.
POTENTIAL APPLICATIONS OF PLANT EVS
The significant role of plant EVs in transporting genetic cargo in plant-pathogen interactions has motivated efforts to develop nanocarrier mimics of these natural EVs for RNAi-based pathogen control strategies in agriculture and as drug delivery agents in the medical field [Figure 3]. These organic nanocarriers can consist of various materials, including lipid-based artificial nanovesicles (AVs) and plant-derived nanovesicles (PDNVs). AVs have recently been used for dsRNA delivery in spray-induced gene silencing (SIGS) to improve the stability of dsRNA in the environment[39,161-163]. SIGS is a powerful and eco-friendly method for crop protection. Many pathogens can efficiently take up RNAs from the environment[88,163], which makes SIGS possible. Topical application of exogenous RNA targeting pest/pathogen virulence genes results in gene silencing and subsequent disease inhibition. AVs formulated with 1,2-dioleoyl 3-trimethylammonium-propane (DOTAP) + polyethylene glycol (PEG), 1,2-dioleyloxy-3-dimethylaminopropane (DODMA), or DOTAP alone were found to provide strong and prolonged protection of dsRNA against B. cinerea on pre- and post-harvest materials[163]. Encapsulation of dsRNA in AVs extended RNAi-based protection for up to 10 days on fruit and 21 days on grape leaves[163]. Similarly, other reports have investigated the use of liposomes and lipid transfection reagents for SIGS against insect pests[164,165].
Figure 3. Potential Application for Plant EVs. Plant-derived EVs and artificial vesicles have been developed for agricultural crop protection and advances in human medicine. Artificial vesicles have been used to load and stabilize pathogen and pest-targeted sRNAs on plants[163], as well as being used in drug delivery mechanisms for human medicine[77,78]. Plant-derived EVs have been explored for their native anticancer, anti-inflammatory, and other medicinal uses in humans, as well as their potential to uptake and deliver drugs through biological barriers within the body. This figure was created with https://www.biorender.com/.
As mentioned above, PDNVs from various fruit and vegetable sources have attracted considerable attention for nanomedicine and drug delivery due to their many favorable properties [Figure 3]. Since PDNVs are derived from plants, they are biocompatible, biodegradable, and easy to scale up. In addition, the small size of PDNVs enhances cellular uptake in humans. PDNVs can also tolerate the gastrointestinal tract and cross biological barriers such as the blood-brain barrier to deliver biological molecules[166,167]. PDNVs may also have unique inherent properties depending on the plant source. For example, PDNVs derived from lemon juice exhibited anticancer activity[168], while ginger EVs possessed anti-inflammatory activity and were preferentially taken up by microbes[75,169]. As well, watermelon-derived extracellular vesicles can influence intestinal cell secretions and consequently, the placental proteome[170]. Thus, PDNVs are gaining interest not only as nanocarriers but also as therapeutic agents themselves that can modulate the activity of microbial and mammalian cells. This has led to significant efforts to characterize the cargo and composition of these PDNVs from various juice sources to better understand their biological functions.
Modifications of PDNVs can make them more suitable for use in therapeutics. For example, lipids can be extracted from PDNVs and reassembled into nanoparticles or treated with chemicals to modify their structure and properties. These types of modifications have been shown to make PDNVs more amendable to the loading of RNAs or specific drugs[171]. More work is needed to fully understand the depth and breadth of PDNVs[166,167]. Finally, PDNVs could have significant agricultural applications by delivering biological cargoes in pathogen control strategies. In particular, the environmentally friendly and scalable nature of PDNVs makes them very attractive compared to current synthetic nanocarriers.
DISCUSSION
The recent emergence and expansion of the studies of plant and microbial EVs has demonstrated the powerful role of EVs in intercellular communication between organisms, taking the field of vesicle biology a new step further beyond the realm of intra-organismal cellular communication. There are evident gaps in the knowledge around plant and microbial EV cargo, biogenesis, and their role in RNA, protein, and metabolite trafficking. New insights into EV-mediated cross-kingdom trafficking are emerging in several host-pathogen/symbiont interactions in both the plant and mammalian fields, with the potential for many more discoveries between species. As we have highlighted, cross-kingdom RNAi is bidirectional, and sRNA trafficking between plants and their interacting organisms induces gene silencing in trans. Cross-kingdom RNAi exists in plant-fungal interactions, as well as between plants and other microbes and pests, including bacteria, oomycetes, and parasites. While nematodes, bacteria, and yeast have been shown to traffic biological material into their mammalian hosts[42,141,143,144], recent studies have shown that mammalian hosts also use EVs to traffic sRNAs to interacting organisms[122].
There is clear evidence that plants use EVs to transport RNA and proteins into interacting organisms, analogous to how mammalian cells use EVs to shuttle RNA and other biological materials between cells or tissues within a single organism or between interacting organisms. Similarly, microbes generate EVs with a variety of contents in order to communicate with other microbes or their hosts. The study of EVs is expanding our understanding of organismal interactions. A new study by Hackl et al. discovered that the marine bacterial species Prochlorococcus secretes vesicles containing strain-specific DNA transposons that facilitate horizontal gene transfer of metabolic and bacteriophage resistance genes between Prochlorococcus strains[172], demonstrating that these EVs are very stable and can survive in seawater. Recently, it was also shown that the algal species Emiliania huxleyi, when infected with E. huxleyi virus, secretes vesicles containing sRNAs that influence the population dynamics of interacting marine microorganisms such as phytoplankton and bacteria[173]. These studies demonstrate the essential role of vesicles in the assembly and communication of marine microbial communities, highlighting the underappreciated role of EVs in inter-organismal cellular interactions, microbial evolution, and environmental stability. More research is needed to understand how microbes such as bacteria and fungi use EVs for adaptation and intra- and inter-species communication.
Although five distinct classes of plant EVs have been identified, the characterization of their biological functions, cargoes, biogenesis and biomarkers lags behind that of mammalian EVs. Furthermore, studies of microbial EVs are even further limited. The functions of cargoes between species are often not as apparent as those between tissues within the same organism. Thus, the cargoes and biological functions of EVs in plants and microbes are and will continue to be an intriguing yet challenging direction in the field of vesicle biology. The current task of the field is to identify more EV markers in plants and microbial species and to improve methods to isolate EVs and purify specific classes of EVs.
Although EV-mediated transport is a key mechanism for RNA protection and delivery between hosts and microbes, EV-independent RNA transport has also been reported to date. In human plasma, extracellular RNA and protein complexes were identified, such as AGO protein-RNA complexes and high-density lipoprotein-RNA complexes[174,175]. In plants, nearly 30% of host Arabidopsis sRNAs found in B. cinerea cells were not found in EVs[29], suggesting an EV-independent pathway for transporting these sRNAs. However, EV-independent extracellular RNAs and RNA-protein complexes would likely undergo rapid degradation in the plant extracellular environment, which contains numerous nucleases and proteases[53,54]. It remains to be explored how EV-independent extracellular RNAs and RNA complexes are secreted and survive in extracellular environments, and whether these RNAs are functional and mediate cross-kingdom RNAi. Overall, encapsulation of RNA in EVs is an effective strategy for cells to protect extracellular RNA from degradation and may also mediate efficient RNA uptake into interacting cells and organisms.
A better understanding of plant EVs and cross-kingdom RNAi will promote the development and application of a new generation of RNA “fungicides” to control plant diseases caused by eukaryotic pathogens[35,163]. In addition, plant-derived EVs and nanovesicles have a potential role in novel medical therapies[77,78]. Research has begun to develop PDNVs into therapeutic agents because of their inherent ability to interact with human cells and influence protein and metabolite composition. However, more research is required to fully characterize the diversity of cargoes and the overall effects of PDNVs. In the future, plant-derived EVs could be engineered to carry specific cargoes, such as therapeutic RNAs or drugs, while providing a protective envelope and effective delivery of the therapeutic agents into human cells.
Gradually, research on plant and microbial EVs will begin to catch up with the greater knowledge of mammalian EVs, and the field of EV-mediated intra-organismal cell-to-cell communication and cross-kingdom/organismal communication will continue to grow. No cell exists alone, and whether as part of the tissues of a larger organism or as a single-celled member of a microbial community, research is increasingly demonstrating the importance of EVs in mediating cargo exchange and communication between cells, tissues, and organisms.
DECLARATIONS
AcknowledgmentWe apologize that we would not be able to include and cite many related interesting studies due to the space limitation.
Authors’ contributionsConceived and designed the structure and content of this review: Jin H, Cai Q
Prepared the initial draft: Cai Q, Halilovic L, He B
Designed and prepared the figures: Jin H, Cai Q, Halilovic L, He B
Collection and collation of literature: Cai Q, Shi T, Halilovic L, Chen A, Wu H
Edited and revised the manuscript: Jin H
Read and approved the final version of the manuscript: Cai Q, Halilovic L, Shi T, Chen A, He B, Wu H, Jin H
Availability of data and materialsNot applicable.
Financial Support and SponsorshipWork in the Cai Q. laboratory was supported by grants from the National Key R&D Program of China (2022YFD1401500); the National Natural Science Foundation of China (32272029, 32070288); Hubei Provincial Natural Science Foundation of China (2022CFA079, 2022CFA025); Work in the H.J. laboratory was supported by the National Institute of Health (R35GM136379); the National Science Foundation (IOS 2020731); United States Department of Agriculture National Institute of Food and Agriculture (2021-67013-34258); United States Department of Agriculture National Institute of Food and Agriculture (2019-70016-29067); And the CIFAR ‘Fungal Kingdom’ fellowship.
Conflicts of interestAll authors declared that there are no conflicts of interest.
Ethical approval and consent to participateNot applicable.
Consent for publicationNot applicable.
Copyright© The Author(s) 2023.
REFERENCES
1. Chargaff E, West R. The biological significance of the thromboplastic protein of blood. J Biol Chem 1946;166:189-97.
2. Yáñez-Mó M, Siljander PR, Andreu Z, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 2015;4:27066.
3. Buzas EI. The roles of extracellular vesicles in the immune system. Nat Rev Immunol 2023;23:236-50.
4. Jensen WA. The composition and ultrastructure of the nucellus in cotton. J Ultrastruct Res 1965;13:112-28.
5. Halperin W, Jensen WA. Ultrastructural changes during growth and embryogenesis in carrot cell cultures. J Ultrastruct Res 1967;18:428-43.
6. An Q, Ehlers K, Kogel KH, van Bel AJ, Hückelhoven R. Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol 2006;172:563-76.
7. Micali CO, Neumann U, Grunewald D, Panstruga R, O'Connell R. Biogenesis of a specialized plant-fungal interface during host cell internalization of golovinomyces orontii haustoria. Cell Microbiol 2011;13:210-26.
8. Wang F, Shang Y, Fan B, Yu JQ, Chen Z. Arabidopsis LIP5, a positive regulator of multivesicular body biogenesis, is a critical target of pathogen-responsive MAPK cascade in plant basal defense. PLoS Pathog 2014;10:e1004243.
9. Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol 2019;21:9-17.
10. Colombo M, Raposo G, Théry C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol 2014;30:255-89.
11. Kim KM, Abdelmohsen K, Mustapic M, Kapogiannis D, Gorospe M. RNA in extracellular vesicles. Wiley Interdiscip Rev RNA 2017;8:e1413.
12. Ghanam J, Chetty VK, Barthel L, Reinhardt D, Hoyer PF, Thakur BK. DNA in extracellular vesicles: from evolution to its current application in health and disease. Cell Biosci 2022;12:37.
13. Casadei L, Sarchet P, de Faria FCC, et al. In situ hybridization to detect DNA amplification in extracellular vesicles. J Extracell Vesicles 2022;11:e12251.
14. O'Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat Rev Mol Cell Biol 2020;21:585-606.
15. Han P, Bartold PM, Salomon C, Ivanovski S. Salivary outer membrane vesicles and dna methylation of small extracellular vesicles as biomarkers for periodontal status: a pilot study. Int J Mol Sci 2021;22:2423.
16. Urabe F, Kosaka N, Ito K, Kimura T, Egawa S, Ochiya T. Extracellular vesicles as biomarkers and therapeutic targets for cancer. Am J Physiol Cell Physiol 2020;318:C29-39.
17. Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science 2020:367.
18. Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol 2018;19:213-28.
19. Sedgwick AE, D'Souza-Schorey C. The biology of extracellular microvesicles. Traffic 2018;19:319-27.
20. Trams EG, Lauter CJ, Salem N Jr, Heine U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim Biophys Acta 1981;645:63-70.
21. Zargarian S, Shlomovitz I, Erlich Z, et al. Phosphatidylserine externalization, "necroptotic bodies" release, and phagocytosis during necroptosis. PLoS Biol 2017;15:e2002711.
22. Yoon S, Kovalenko A, Bogdanov K, Wallach D. MLKL, the protein that mediates necroptosis, also regulates endosomal trafficking and extracellular vesicle generation. Immunity 2017;47:51-65.e7.
24. Hristov M, Erl W, Linder S, Weber PC. Apoptotic bodies from endothelial cells enhance the number and initiate the differentiation of human endothelial progenitor cells in vitro. Blood 2004;104:2761-6.
25. Cheng L, Hill AF. Therapeutically harnessing extracellular vesicles. Nat Rev Drug Discov 2022;21:379-99.
26. Wiklander OPB, Brennan MÁ, Lötvall J, Breakefield XO, El Andaloussi S. Advances in therapeutic applications of extracellular vesicles. Sci Transl Med 2019;11.
27. Urzì O, Raimondo S, Alessandro R. Extracellular vesicles from plants: current knowledge and open questions. Int J Mol Sci 2021;22:5366.
28. Vojtech L, Woo S, Hughes S, et al. Exosomes in human semen carry a distinctive repertoire of small non-coding RNAs with potential regulatory functions. Nucleic Acids Res 2014;42:7290-304.
29. Cai Q, Qiao L, Wang M, et al. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018;360:1126-9.
30. Huang Y, Wang S, Cai Q, Jin H. Effective methods for isolation and purification of extracellular vesicles from plants. J Integr Plant Biol 2021;63:2020-30.
31. Rutter BD, Innes RW. Extracellular vesicles isolated from the leaf apoplast carry stress-response proteins. Plant Physiol 2017;173:728-41.
32. Regente M, Corti-Monzón G, Maldonado AM, et al. Vesicular fractions of sunflower apoplastic fluids are associated with potential exosome marker proteins. FEBS Lett 2009;583:3363-6.
33. Boccia E, Alfieri M, Belvedere R, et al. Plant hairy roots for the production of extracellular vesicles with antitumor bioactivity. Commun Biol 2022;5:848.
34. De Palma M, Ambrosone A, Leone A, et al. Plant roots release small extracellular vesicles with antifungal activity. Plants 2020;9:1777.
35. Cai Q, He B, Weiberg A, Buck AH, Jin H. Small RNAs and extracellular vesicles: new mechanisms of cross-species communication and innovative tools for disease control. PLoS Pathog 2019;15:e1008090.
36. Cai Q, He B, Jin H. A safe ride in extracellular vesicles - small RNA trafficking between plant hosts and pathogens. Curr Opin Plant Biol 2019;52:140-8.
37. Huang CY, Wang H, Hu P, Hamby R, Jin H. Small RNAs - big players in plant-microbe interactions. Cell Host Microbe 2019;26:173-82.
38. Cai Q, He B, Kogel KH, Jin H. Cross-kingdom RNA trafficking and environmental RNAi-nature's blueprint for modern crop protection strategies. Curr Opin Microbiol 2018;46:58-64.
39. Wang M, Thomas N, Jin H. Cross-kingdom RNA trafficking and environmental RNAi for powerful innovative pre- and post-harvest plant protection. Curr Opin Plant Biol 2017;38:133-41.
40. Knip M, Constantin ME, Thordal-Christensen H. Trans-kingdom cross-talk: small RNAs on the move. Plos Genetics 2014;10:e1004602.
41. Wang M, Dean RA. Movement of small RNAs in and between plants and fungi. Mol Plant Pathol 2020;21:589-601.
42. Buck AH, Coakley G, Simbari F, et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat Commun 2014;5:5488.
43. Hou Y, Zhai Y, Feng L, et al. A phytophthora effector suppresses trans-kingdom RNAi to promote disease susceptibility. Cell Host Microbe 2019;25:153-165.e5.
44. Liu S, da Cunha AP, Rezende RM, et al. The host shapes the gut microbiota via fecal microRNA. Cell Host Microbe 2016;19:32-43.
45. Cai Q, He B, Wang S, et al. Message in a bubble: shuttling small rnas and proteins between cells and interacting organisms using extracellular vesicles. Annu Rev Plant Biol 2021;72:497-524.
46. Choi JW, Kim SC, Hong SH, Lee HJ. Secretable small rnas via outer membrane vesicles in periodontal pathogens. J Dent Res 2017;96:458-66.
47. Costa JH, Bazioli JM, Barbosa LD, et al. Phytotoxic tryptoquialanines produced
48. Wang Z, Zeng J, Deng J, et al. Pathogen-derived extracellular vesicles: emerging mediators of plant-microbe interactions. Mol Plant Microbe Interact 2023;36:218-27.
49. Garcia-Ceron D, Lowe RGT, McKenna JA, et al. Extracellular vesicles from fusarium graminearum contain protein effectors expressed during infection of corn. J Fungi 2021;7:977.
50. Hill EH, Solomon PS. Extracellular vesicles from the apoplastic fungal wheat pathogen Zymoseptoria tritici. Fungal Biol Biotechnol 2020;7:13.
51. Bleackley MR, Samuel M, Garcia-Ceron D, et al. Extracellular vesicles from the cotton pathogen fusarium oxysporum f. sp. vasinfectum induce a phytotoxic response in plants. Front Plant Sci 2019;10:1610.
52. Fang Y, Wang Z, Zhang S, Peng Q, Liu X. Characterization and proteome analysis of the extracellular vesicles of phytophthora capsici. J Proteomics 2021;238:104137.
53. Wang Y, Wang Y, Wang Y. Apoplastic proteases: powerful weapons against pathogen infection in plants. Plant Commun 2020;1:100085.
54. Delaunois B, Colby T, Belloy N, et al. Large-scale proteomic analysis of the grapevine leaf apoplastic fluid reveals mainly stress-related proteins and cell wall modifying enzymes. BMC Plant Biol 2013;13:24.
55. Roth R, Hillmer S, Funaya C, et al. Arbuscular cell invasion coincides with extracellular vesicles and membrane tubules. Nat Plants 2019;5:204-11.
56. He B, Cai Q, Qiao L, et al. RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles. Nat Plants 2021;7:342-52.
57. Regente M, Pinedo M, San Clemente H, Balliau T, Jamet E, de la Canal L. Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. J Exp Bot 2017;68:5485-95.
58. Prado N, De Linares C, Sanz ML, et al. Pollensomes as natural vehicles for pollen allergens. J Immunol 2015;195:445-9.
59. Prado N, Alché Jde D, Casado-Vela J, et al. Nanovesicles are secreted during pollen germination and pollen tube growth: a possible role in fertilization. Mol Plant 2014;7:573-7.
60. Movahed N, Cabanillas DG, Wan J, Vali H, Laliberté JF, Zheng H. Turnip mosaic virus components are released into the extracellular space by vesicles in infected leaves. Plant Physiol 2019;180:1375-88.
61. Kowal J, Arras G, Colombo M, et al. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc Natl Acad Sci USA 2016;113:E968-77.
62. Gurunathan S, Kang MH, Qasim M, Khan K, Kim JH. Biogenesis, membrane trafficking, functions, and next generation nanotherapeutics medicine of extracellular vesicles. Int J Nanomedicine 2021;16:3357-83.
63. Baldrich P, Rutter BD, Karimi HZ, Podicheti R, Meyers BC, Innes RW. Plant extracellular vesicles contain diverse small RNA species and are enriched in 10- to 17-nucleotide "tiny" RNAs. Plant Cell 2019;31:315-24.
64. Jeon HS, Jang E, Kim J, et al. Pathogen-induced autophagy regulates monolignol transport and lignin formation in plant immunity. Autophagy 2023;19:597-615.
65. Doyle LM, Wang MZ. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 2019;8:727.
66. Trajkovic K, Hsu C, Chiantia S, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008;319:1244-7.
67. Liu NJ, Wang N, Bao JJ, Zhu HX, Wang LJ, Chen XY. Lipidomic analysis reveals the importance of GIPCs in arabidopsis leaf extracellular vesicles. Mol Plant 2020;13:1523-32.
68. Gronnier J, Germain V, Gouguet P, Cacas JL, Mongrand S. GIPC: glycosyl inositol phospho ceramides, the major sphingolipids on earth. Plant Signal Behav 2016;11:e1152438.
69. Kwon C, Neu C, Pajonk S, et al. Co-option of a default secretory pathway for plant immune responses. Nature 2008;451:835-40.
70. Ding Y, Wang J, Chun Lai JH, et al. Exo70E2 is essential for exocyst subunit recruitment and EXPO formation in both plants and animals. Mol Biol Cell 2014;25:412-26.
71. Wang J, Ding Y, Wang J, et al. EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell 2010;22:4009-30.
72. Kameli N, Dragojlovic-Kerkache A, Savelkoul P, Stassen FR. Plant-derived extracellular vesicles: current findings, challenges, and future applications. Membranes 2021;11:411.
73. Zhang HG, Cao P, Teng Y, et al. Isolation, identification, and characterization of novel nanovesicles. Oncotarget 2016;7:41346-62.
74. Wang QL, Zhuang XY, Mu JY, et al. Delivery of therapeutic agents by nanoparticles made of grapefruit-derived lipids. Nat Commun 2013;4:1867.
75. Teng Y, Ren Y, Sayed M, et al. Plant-derived exosomal microRNAs shape the gut microbiota. Cell Host Microbe 2018;24:637-52.
76. Liu Y, Wu S, Koo Y, et al. Characterization of and isolation methods for plant leaf nanovesicles and small extracellular vesicles. Nanomedicine 2020;29:102271.
77. Tan ZL, Li JF, Luo HM, Liu YY, Jin Y. Plant extracellular vesicles: a novel bioactive nanoparticle for tumor therapy. Front Pharmacol 2022;13:1006299.
78. Karamanidou T, Tsouknidas A. Plant-derived extracellular vesicles as therapeutic nanocarriers. Int J Mol Sci 2022;23:191.
79. Zand Karimi H, Baldrich P, Rutter BD, et al. Arabidopsis apoplastic fluid contains sRNA- and circular RNA-protein complexes that are located outside extracellular vesicles. Plant Cell 2022;34:1863-81.
80. Chen A, He B, Jin H. Isolation of extracellular vesicles from arabidopsis. Curr Protoc 2022;2:e352.
81. Wubbolts R, Leckie RS, Veenhuizen PT, et al. Proteomic and biochemical analyses of human B cell-derived exosomes. potential implications for their function and multivesicular body formation. J Biol Chem 2003;278:10963-72.
82. Jeppesen DK, Fenix AM, Franklin JL, et al. Reassessment of exosome composition. Cell 2019;177:428-445.e18.
83. Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol 2015;25:364-72.
84. Kim YB, Lee GB, Moon MH. Size separation of exosomes and microvesicles using flow field-flow fractionation/multiangle light scattering and lipidomic comparison. Anal Chem 2022;94:8958-65.
85. Katiyar-Agarwal S, Jin H. Role of small RNAs in host-microbe interactions. Annu Rev Phytopathol 2010;48:225-46.
86. Lopez-Gomollon S, Baulcombe DC. Roles of RNA silencing in viral and non-viral plant immunity and in the crosstalk between disease resistance systems. Nat Rev Mol Cell Biol 2022;23:645-62.
87. Rosa C, Kuo YW, Wuriyanghan H, Falk BW. RNA interference mechanisms and applications in plant pathology. Annu Rev Phytopathol 2018;56:581-610.
88. Wang M, Weiberg A, Lin FM, Thomma BP, Huang HD, Jin H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat Plants 2016;2:16151.
89. Weiberg A, Wang M, Bellinger M, Jin H. Small RNAs: a new paradigm in plant-microbe interactions. Annu Rev Phytopathol 2014;52:495-516.
90. Chen X, Rechavi O. Plant and animal small RNA communications between cells and organisms. Nat Rev Mol Cell Biol 2022;23:185-203.
91. Weiberg A, Wang M, Lin FM, et al. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013;342:118-23.
92. He B, Cai Q, Weiberg A, et al. Botrytis cinerea small RNAs are associated with tomato AGO1 and silence tomato defense-related target genes supporting cross-kingdom RNAi. bioRxiv :2023.
93. Tang D, Wang G, Zhou JM. Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell 2017;29:618-37.
94. Zhang BS, Li YC, Guo HS, Zhao JH. Verticillium dahliae secretes small RNA to target host MIR157d and retard plant floral transition during infection. Front Plant Sci 2022;13:847086.
95. Wang B, Sun Y, Song N, et al. Puccinia striiformis f. sp. tritici microRNA-like RNA 1 (Pst-milR1), an important pathogenicity factor of Pst, impairs wheat resistance to Pst by suppressing the wheat pathogenesis-related 2 gene. New Phytol 2017;215:338-50.
96. Mueth NA, Ramachandran SR, Hulbert SH. Small RNAs from the wheat stripe rust fungus (Puccinia striiformis f.sp. tritici). BMC Genomics 2015;16:718.
97. Ji HM, Mao HY, Li SJ, et al. Fol-milR1, a pathogenicity factor of Fusarium oxysporum, confers tomato wilt disease resistance by impairing host immune responses. New Phytol 2021;232:705-18.
98. Dunker F, Trutzenberg A, Rothenpieler JS, et al. Oomycete small RNAs bind to the plant RNA-induced silencing complex for virulence. Elife 2020:9.
99. Kusch S, Singh M, Thieron H, Spanu PD, Panstruga R. Site-specific analysis reveals candidate cross-kingdom small RNAs, tRNA and rRNA fragments, and signs of fungal RNA phasing in the barley-powdery mildew interaction. Mol Plant Pathol 2023;24:570-87.
100. Duanis-Assaf D, Galsurker O, Davydov O, et al. Double-stranded RNA targeting fungal ergosterol biosynthesis pathway controls Botrytis cinerea and postharvest grey mould. Plant Biotechnol J 2022;20:226-37.
101. Werner BT, Koch A, Šečić E, et al. Fusarium graminearum DICER-like-dependent sRNAs are required for the suppression of host immune genes and full virulence. PLoS One 2021;16:e0252365.
102. Yin C, Zhu H, Jiang Y, Shan Y, Gong L. Silencing dicer-like genes reduces virulence and sRNA generation in penicillium italicum, the cause of citrus blue mold. Cells 2020;9:363.
103. Feng H, Xu M, Liu Y, Dong R, Gao X, Huang L. Dicer-like genes are required for H(2)O(2) and KCl stress responses, pathogenicity and small RNA generation in valsa mali. Front Microbiol 2017;8:1166.
104. Haile ZM, Gebremichael DE, Capriotti L, et al. Double-stranded RNA targeting dicer-like genes compromises the pathogenicity of plasmopara viticola on grapevine. Front Plant Sci 2021;12:667539.
105. Islam MT, Davis Z, Chen L, et al. Minicell-based fungal RNAi delivery for sustainable crop protection. Microb Biotechnol 2021;14:1847-56.
106. Werner BT, Gaffar FY, Schuemann J, Biedenkopf D, Koch AM. RNA-spray-mediated silencing of fusarium graminearum AGO and DCL genes improve barley disease resistance. Front Plant Sci 2020;11:476.
107. Wang Q, An B, Hou X, Guo Y, Luo H, He C. Dicer-like proteins regulate the growth, conidiation, and pathogenicity of colletotrichum gloeosporioides from hevea brasiliensis. Front Microbiol 2017;8:2621.
108. Hollister JD, Smith LM, Guo YL, Ott F, Weigel D, Gaut BS. Transposable elements and small RNAs contribute to gene expression divergence between Arabidopsis thaliana and Arabidopsis lyrata. Proc Natl Acad Sci USA 2011;108:2322-7.
109. Martinez F, Dubos B, Fermaud M. The role of saprotrophy and virulence in the population dynamics of botrytis cinerea in vineyards. Phytopathology 2005;95:692-700.
110. Shahid S, Kim G, Johnson NR, et al. MicroRNAs from the parasitic plant cuscuta campestris target host messenger RNAs. Nature 2018;553:82-5.
111. Wong-Bajracharya J, Singan VR, Monti R, et al. The ectomycorrhizal fungus Pisolithus microcarpus encodes a microRNA involved in cross-kingdom gene silencing during symbiosis. Proc Natl Acad Sci USA 2022:119.
112. Silvestri A, Fiorilli V, Miozzi L, Accotto GP, Turina M, Lanfranco L. In silico analysis of fungal small RNA accumulation reveals putative plant mRNA targets in the symbiosis between an arbuscular mycorrhizal fungus and its host plant. BMC Genomics 2019;20:169.
113. Ren B, Wang X, Duan J, Ma J. Rhizobial tRNA-derived small RNAs are signal molecules regulating plant nodulation. Science 2019;365:919-22.
114. Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through dicer-like 1 protein functions. Proc Natl Acad Sci U S A 2004;101:12753-8.
115. Meng X, Jin W, Wu F. Novel tomato miRNA miR1001 initiates cross-species regulation to suppress the conidiospore germination and infection virulence of Botrytis cinerea in vitro. Gene 2020;759:145002.
116. Wu F, Huang Y, Jiang W, Jin W. Genome-wide identification and validation of tomato-encoded sRNA as the cross-species antifungal factors targeting the virulence genes of Botrytis cinerea. Front Plant Sci 2023;14:1072181.
117. Zhang T, Zhao YL, Zhao JH, et al. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat Plants 2016;2:16153.
118. Jiao J, Peng D. Wheat microRNA1023 suppresses invasion of
119. Zhu C, Liu JH, Zhao JH, et al. A fungal effector suppresses the nuclear export of AGO1-miRNA complex to promote infection in plants. Proc Natl Acad Sci USA 2022;119:e2114583119.
120. Cui C, Wang Y, Liu J, Zhao J, Sun P, Wang S. A fungal pathogen deploys a small silencing RNA that attenuates mosquito immunity and facilitates infection. Nat Commun 2019;10:4298.
121. Wang Y, Cui C, Wang G, Li Y, Wang S. Insects defend against fungal infection by employing microRNAs to silence virulence-related genes. Proc Natl Acad Sci USA 2021:118.
122. Halder LD, Babych S, Palme DI, et al. Candida albicans Induces cross-kingdom miRNA trafficking in human monocytes to promote fungal growth. mBio 2021;13:e0356321.
123. Hirschi KD, Pruss GJ, Vance V. Dietary delivery: a new avenue for microRNA therapeutics? Trends Biotechnol 2015;33:431-2.
124. Melo SA, Sugimoto H, O'Connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 2014;26:707-21.
125. Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2011;2:282.
126. Qu L, Ding J, Chen C, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell 2016;29:653-68.
127. Yokoi A, Yoshioka Y, Yamamoto Y, et al. Malignant extracellular vesicles carrying MMP1 mRNA facilitate peritoneal dissemination in ovarian cancer. Nat Commun 2017;8:14470.
128. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654-9.
129. Ratajczak J, Miekus K, Kucia M, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 2006;20:847-56.
130. Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Cabo F, et al. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 2013;4:2980.
131. Wozniak AL, Adams A, King KE, et al. The RNA binding protein FMR1 controls selective exosomal miRNA cargo loading during inflammation. J Cell Biol 2020:219.
132. Luo X, Jean-Toussaint R, Sacan A, Ajit SK. Differential RNA packaging into small extracellular vesicles by neurons and astrocytes. Cell Commun Signal 2021;19:75.
133. Chow FW, Koutsovoulos G, Ovando-Vázquez C, et al. Secretion of an argonaute protein by a parasitic nematode and the evolution of its siRNA guides. Nucleic Acids Res 2019;47:3594-606.
134. Brown L, Wolf JM, Prados-Rosales R, Casadevall A. Through the wall: extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat Rev Microbiol 2015;13:620-30.
135. de Toledo Martins S, Szwarc P, Goldenberg S, Alves LR. Extracellular vesicles in fungi: composition and functions. In: Rodrigues ML, editor. Fungal Physiology and Immunopathogenesis. Cham: Springer International Publishing; 2019. pp. 45-59.
136. Caruana JC, Walper SA. Bacterial membrane vesicles as mediators of microbe - microbe and microbe - host community interactions. Front Microbiol 2020;11:432.
137. Badi S, Bruno SP, Moshiri A, Tarashi S, Siadat SD, Masotti A. Small RNAs in outer membrane vesicles and their function in host-microbe interactions. Front Microbiol 2020;11:1209.
138. Cecil JD, Sirisaengtaksin N, O'Brien-Simpson NM, Krachler AM. Outer membrane vesicle-host cell interactions. Microbiol Spectr 2019;7:10.1128;.
139. Gow NAR, Lenardon MD. Architecture of the dynamic fungal cell wall. Nat Rev Microbiol 2023;21:248-59.
140. Wolf JM, Espadas-Moreno J, Luque-Garcia JL, Casadevall A. Interaction of Cryptococcus neoformans extracellular vesicles with the cell wall. Eukaryot Cell 2014;13:1484-93.
141. Liebana-Jordan M, Brotons B, Falcon-Perez JM, Gonzalez E. Extracellular vesicles in the fungi kingdom. Int J Mol Sci 2021;22:7221.
142. Rizzo J, Rodrigues ML, Janbon G. Extracellular vesicles in fungi: past, present, and future perspectives. Front Cell Infect Microbiol 2020;10:346.
143. Janda M, Robatzek S. Extracellular vesicles from phytobacteria: Properties, functions and uses. Biotechnol Adv 2022;58:107934.
144. Chronopoulos A, Kalluri R. Emerging role of bacterial extracellular vesicles in cancer. Oncogene 2020;39:6951-60.
145. Rodrigues ML, Nimrichter L, Oliveira DL, et al. Vesicular polysaccharide export in Cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell 2007;6:48-59.
146. Zarnowski R, Sanchez H, Jaromin A, et al. A common vesicle proteome drives fungal biofilm development. Proc Natl Acad Sci U S A 2022;119:e2211424119.
147. Bahar O, Pruitt R, Luu DD, et al. The Xanthomonas Ax21 protein is processed by the general secretory system and is secreted in association with outer membrane vesicles. PeerJ 2014;2:e242.
148. Sidhu VK, Vorhölter FJ, Niehaus K, Watt SA. Analysis of outer membrane vesicle associated proteins isolated from the plant pathogenic bacterium Xanthomonas campestris pv. campestris. BMC Microbiol 2008;8:87.
149. Solé M, Scheibner F, Hoffmeister AK, et al. Xanthomonas campestris pv. vesicatoria Secretes Proteases and Xylanases via the Xps Type II Secretion System and Outer Membrane Vesicles. J Bacteriol 2015;197:2879-93.
150. Tran TM, Chng CP, Pu X, et al. Potentiation of plant defense by bacterial outer membrane vesicles is mediated by membrane nanodomains. Plant Cell 2022;34:395-417.
151. Feitosa-Junior OR, Stefanello E, Zaini PA, et al. Proteomic and metabolomic analyses of xylella fastidiosa OMV-enriched fractions reveal association with virulence factors and signaling molecules of the DSF family. Phytopathology 2019;109:1344-53.
152. Ionescu M, Zaini PA, Baccari C, Tran S, da Silva AM, Lindow SE. Xylella fastidiosa outer membrane vesicles modulate plant colonization by blocking attachment to surfaces. Proc Natl Acad Sci USA 2014;111:E3910-8.
153. Chowdhury C, Jagannadham MV. Virulence factors are released in association with outer membrane vesicles of Pseudomonas syringae pv. tomato T1 during normal growth. Biochim Biophys Acta 2013;1834:231-9.
154. Janda MCL, Rybak K, Meng C, et al. Biophysical and proteomic analyses suggest functions of Pseudomonas syringae pv tomato DC3000 extracellular vesicles in bacterial growth during plant infection. BioRxiv :2021.
155. McMillan HM, Zebell SG, Ristaino JB, Dong X, Kuehn MJ. Protective plant immune responses are elicited by bacterial outer membrane vesicles. Cell Rep 2021;34:108645.
156. Jäger J, Keese S, Roessle M, Steinert M, Schromm AB. Fusion of Legionella pneumophila outer membrane vesicles with eukaryotic membrane systems is a mechanism to deliver pathogen factors to host cell membranes. Cell Microbiol 2015;17:607-20.
157. Bomberger JM, Maceachran DP, Coutermarsh BA, Ye S, O'Toole GA, Stanton BA. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog 2009;5:e1000382.
158. Ludwig N, Reissmann S, Schipper K, et al. A cell surface-exposed protein complex with an essential virulence function in Ustilago maydis. Nat Microbiol 2021;6:722-30.
159. Kwon S, Rupp O, Brachmann A, et al. mRNA Inventory of Extracellular Vesicles from Ustilago maydis. J Fungi 2021;7:562.
160. Rutter BD, Chu TT, Dallery JF, Zajt KK, O'Connell RJ, Innes RW. The development of extracellular vesicle markers for the fungal phytopathogen colletotrichum higginsianum. J Extracell Vesicles 2022;11:e12216.
161. Wytinck N, Manchur CL, Li VH, Whyard S, Belmonte MF. dsRNA uptake in plant pests and pathogens: insights into RNAi-based insect and fungal control technology. Plants 2020;9:1780.
162. Rank AP, Koch A. Lab-to-field transition of RNA spray applications - how far are we? Front Plant Sci 2021;12:755203.
163. Qiao L, Niño-Sánchez J, Hamby R, et al. Artificial nanovesicles for dsRNA delivery in spray-induced gene silencing for crop protection. Plant Biotechnol J 2023;21:854-65.
164. Castellanos NL, Smagghe G, Sharma R, Oliveira EE, Christiaens O. Liposome encapsulation and EDTA formulation of dsRNA targeting essential genes increase oral RNAi-caused mortality in the Neotropical stink bug Euschistus heros. Pest Manag Sci 2019;75:537-48.
165. Dhandapani RK, Gurusamy D, Palli SR. Protamine-lipid-dsRNA nanoparticles improve RNAi efficiency in the fall armyworm, spodoptera frugiperda. J Agric Food Chem 2022;70:6634-43.
166. Gioia S, Hossain MN, Conese M. Biological properties and therapeutic effects of plant-derived nanovesicles. Open Med 2020;15:1096-122.
167. Dad HA, Gu TW, Zhu AQ, Huang LQ, Peng LH. Plant exosome-like nanovesicles: emerging therapeutics and drug delivery nanoplatforms. Mol Ther 2021;29:13-31.
168. Raimondo S, Naselli F, Fontana S, et al. Citrus limon-derived nanovesicles inhibit cancer cell proliferation and suppress CML xenograft growth by inducing TRAIL-mediated cell death. Oncotarget 2015;6:19514-27.
169. Chen X, Zhou Y, Yu J. Exosome-like nanoparticles from ginger rhizomes inhibited NLRP3 inflammasome activation. Mol Pharm 2019;16:2690-9.
170. Timms K, Holder B, Day A, Mclaughlin J, Forbes KA, Westwood M. Watermelon-derived extracellular vesicles influence human EX vivo placental cell behavior by altering intestinal secretions. Mol Nutr Food Res 2022;66:e2200013.
171. Cong M, Tan S, Li S, et al. Technology insight: Plant-derived vesicles-how far from the clinical biotherapeutics and therapeutic drug carriers? Adv Drug Deliv Rev 2022;182:114108.
172. Hackl T, Laurenceau R, Ankenbrand MJ, et al. Novel integrative elements and genomic plasticity in ocean ecosystems. Cell 2023;186:47-62.e16.
173. Schatz D, Schleyer G, Saltvedt MR, Sandaa RA, Feldmesser E, Vardi A. Ecological significance of extracellular vesicles in modulating host-virus interactions during algal blooms. ISME J 2021;15:3714-21.
174. Arroyo JD, Chevillet JR, Kroh EM, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A 2011;108:5003-8.
Cite This Article
Export citation file: BibTeX | RIS
OAE Style
Cai Q, Halilovic L, Shi T, Chen A, He B, Wu H, Jin H. Extracellular vesicles: cross-organismal RNA trafficking in plants, microbes, and mammalian cells. Extracell Vesicles Circ Nucleic Acids 2023;4:262-82. http://dx.doi.org/10.20517/evcna.2023.10
AMA Style
Cai Q, Halilovic L, Shi T, Chen A, He B, Wu H, Jin H. Extracellular vesicles: cross-organismal RNA trafficking in plants, microbes, and mammalian cells. Extracellular Vesicles and Circulating Nucleic Acids. 2023; 4(2): 262-82. http://dx.doi.org/10.20517/evcna.2023.10
Chicago/Turabian Style
Cai, Qiang, Lida Halilovic, Ting Shi, Angela Chen, Baoye He, Huaitong Wu, Hailing Jin. 2023. "Extracellular vesicles: cross-organismal RNA trafficking in plants, microbes, and mammalian cells" Extracellular Vesicles and Circulating Nucleic Acids. 4, no.2: 262-82. http://dx.doi.org/10.20517/evcna.2023.10
ACS Style
Cai, Q.; Halilovic L.; Shi T.; Chen A.; He B.; Wu H.; Jin H. Extracellular vesicles: cross-organismal RNA trafficking in plants, microbes, and mammalian cells. Extracell. Vesicles. Circ. Nucleic. Acids. 2023, 4, 262-82. http://dx.doi.org/10.20517/evcna.2023.10
About This Article
Special Issue
Copyright
Data & Comments
Data
 Cite This Article 24 clicks
Cite This Article 24 clicks




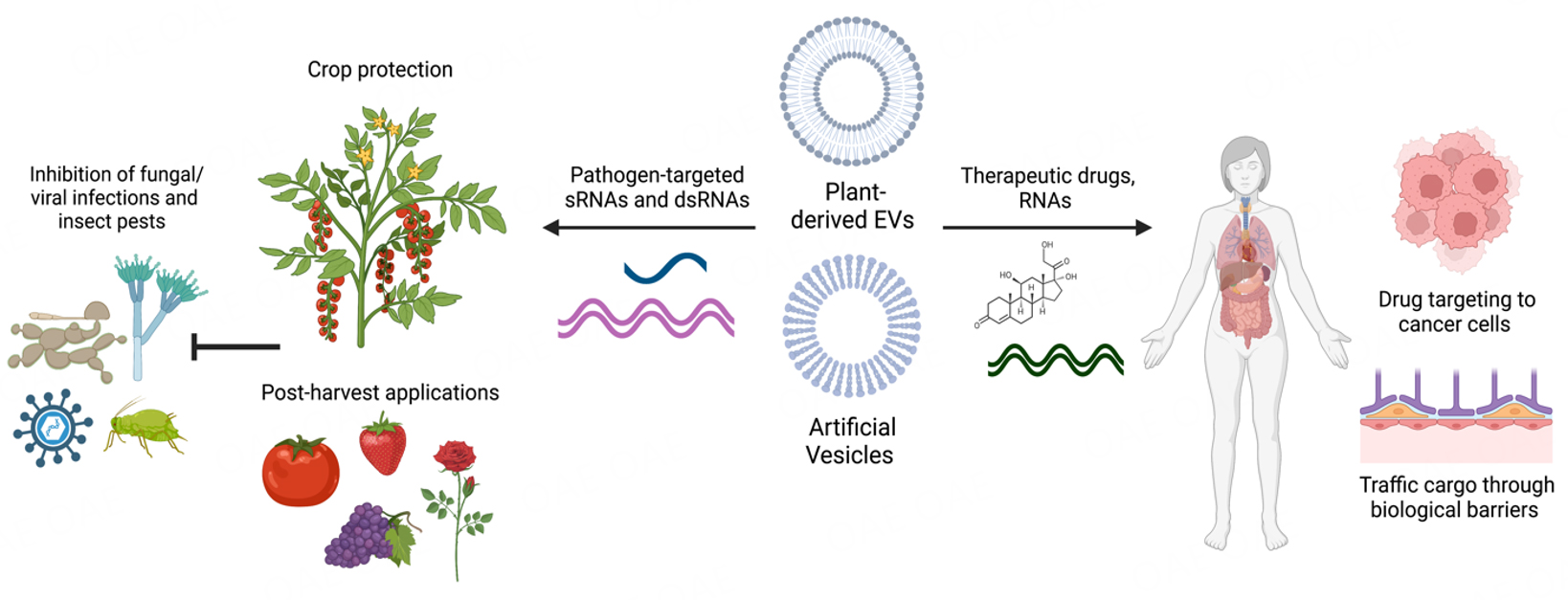










Comments
Comments must be written in English. Spam, offensive content, impersonation, and private information will not be permitted. If any comment is reported and identified as inappropriate content by OAE staff, the comment will be removed without notice. If you have any queries or need any help, please contact us at support@oaepublish.com.